| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
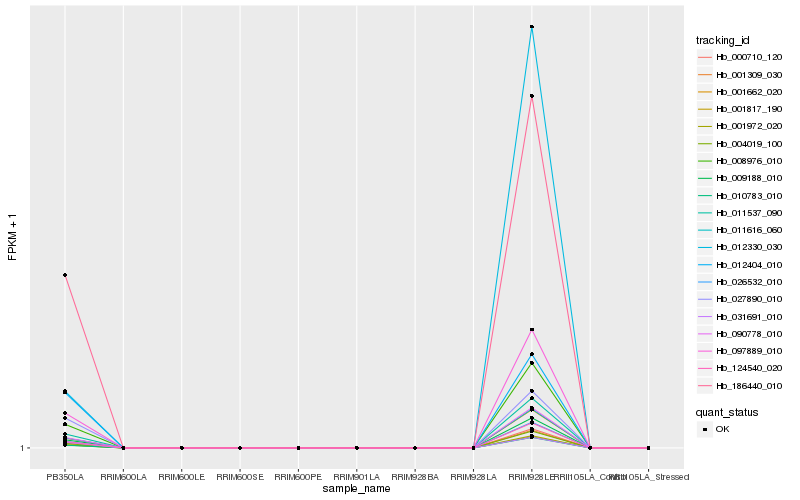
Hb_026532_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 2 |
Hb_001662_020 |
0.0021470397 |
- |
- |
PREDICTED: F-box protein SKIP23-like isoform X3 [Eucalyptus grandis] |
| 3 |
Hb_011616_060 |
0.0022468702 |
- |
- |
PREDICTED: uncharacterized protein LOC105789623 [Gossypium raimondii] |
| 4 |
Hb_090778_010 |
0.0024772239 |
- |
- |
PREDICTED: uncharacterized protein LOC105764482 [Gossypium raimondii] |
| 5 |
Hb_124540_020 |
0.0036573265 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 6 |
Hb_010783_010 |
0.0089689326 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_001817_190 |
0.0090805336 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 8 |
Hb_001972_020 |
0.0092520282 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_004019_100 |
0.0100204608 |
- |
- |
PREDICTED: uncharacterized protein LOC105797448 [Gossypium raimondii] |
| 10 |
Hb_001309_030 |
0.0101621667 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_000710_120 |
0.0104269774 |
- |
- |
hypothetical protein JCGZ_01844 [Jatropha curcas] |
| 12 |
Hb_008976_010 |
0.0108312907 |
- |
- |
- |
| 13 |
Hb_009188_010 |
0.0122936129 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 14 |
Hb_011537_090 |
0.0158957899 |
- |
- |
hypothetical protein B456_009G071100 [Gossypium raimondii] |
| 15 |
Hb_097889_010 |
0.0177918813 |
- |
- |
PREDICTED: uncharacterized protein LOC102584707 [Solanum tuberosum] |
| 16 |
Hb_186440_010 |
0.0965963482 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_012330_030 |
0.1328879212 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_027890_010 |
0.1347611443 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 19 |
Hb_031691_010 |
0.1388410615 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_012404_010 |
0.1568542051 |
- |
- |
- |