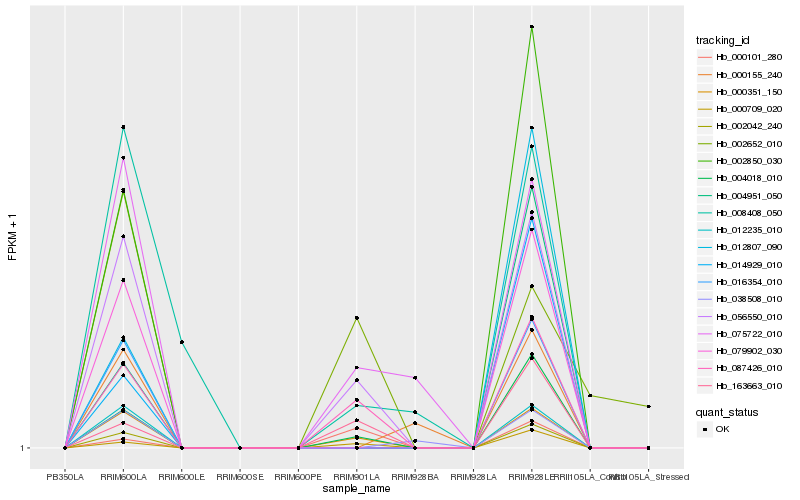
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000351_150 |
0.0 |
- |
- |
Wall-associated kinase 2, putative [Theobroma cacao] |
| 2 |
Hb_056550_010 |
0.0123227552 |
- |
- |
PREDICTED: uncharacterized protein LOC104112577 [Nicotiana tomentosiformis] |
| 3 |
Hb_087426_010 |
0.1056045046 |
- |
- |
PREDICTED: uncharacterized protein LOC105803348 [Gossypium raimondii] |
| 4 |
Hb_004018_010 |
0.1717881243 |
- |
- |
PREDICTED: uncharacterized protein LOC105784835 [Gossypium raimondii] |
| 5 |
Hb_000709_020 |
0.1994153475 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_075722_010 |
0.2206513514 |
- |
- |
hypothetical protein JCGZ_01984 [Jatropha curcas] |
| 7 |
Hb_163663_010 |
0.2366366804 |
- |
- |
PREDICTED: uncharacterized protein LOC105629934 [Jatropha curcas] |
| 8 |
Hb_012235_010 |
0.2466495534 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 9 |
Hb_016354_010 |
0.2471056807 |
- |
- |
- |
| 10 |
Hb_079902_030 |
0.24967873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002042_240 |
0.2563086195 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_002850_030 |
0.2646607342 |
- |
- |
- |
| 13 |
Hb_000101_280 |
0.3021738324 |
- |
- |
hypothetical protein POPTR_0018s02450g [Populus trichocarpa] |
| 14 |
Hb_008408_050 |
0.3078138303 |
- |
- |
- |
| 15 |
Hb_002652_010 |
0.3159185642 |
- |
- |
- |
| 16 |
Hb_038508_010 |
0.3188007816 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 17 |
Hb_012807_090 |
0.3219674532 |
- |
- |
PREDICTED: uncharacterized protein LOC100267173 [Vitis vinifera] |
| 18 |
Hb_004951_050 |
0.3267686911 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000155_240 |
0.3290977003 |
- |
- |
PREDICTED: uncharacterized protein LOC104888642 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_014929_010 |
0.3291411106 |
- |
- |
- |