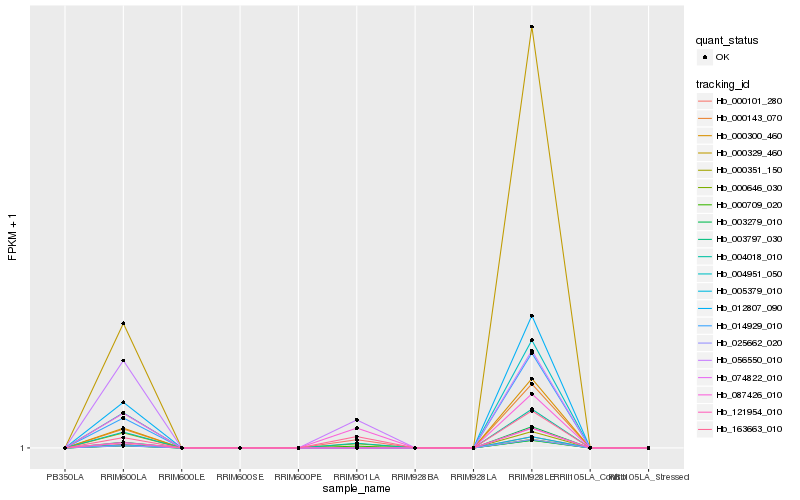
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000709_020 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_163663_010 |
0.0517028527 |
- |
- |
PREDICTED: uncharacterized protein LOC105629934 [Jatropha curcas] |
| 3 |
Hb_004018_010 |
0.107592817 |
- |
- |
PREDICTED: uncharacterized protein LOC105784835 [Gossypium raimondii] |
| 4 |
Hb_087426_010 |
0.1223749122 |
- |
- |
PREDICTED: uncharacterized protein LOC105803348 [Gossypium raimondii] |
| 5 |
Hb_056550_010 |
0.1904081283 |
- |
- |
PREDICTED: uncharacterized protein LOC104112577 [Nicotiana tomentosiformis] |
| 6 |
Hb_000351_150 |
0.1994153475 |
- |
- |
Wall-associated kinase 2, putative [Theobroma cacao] |
| 7 |
Hb_000101_280 |
0.2004662622 |
- |
- |
hypothetical protein POPTR_0018s02450g [Populus trichocarpa] |
| 8 |
Hb_012807_090 |
0.2836527466 |
- |
- |
PREDICTED: uncharacterized protein LOC100267173 [Vitis vinifera] |
| 9 |
Hb_004951_050 |
0.2838726459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_014929_010 |
0.2840502206 |
- |
- |
- |
| 11 |
Hb_000143_070 |
0.2845982928 |
- |
- |
- |
| 12 |
Hb_000300_460 |
0.285204467 |
- |
- |
Mavicyanin, putative [Ricinus communis] |
| 13 |
Hb_003279_010 |
0.2855470025 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 14 |
Hb_121954_010 |
0.2855772383 |
- |
- |
PREDICTED: uncharacterized protein LOC105774125 [Gossypium raimondii] |
| 15 |
Hb_074822_010 |
0.2855859244 |
- |
- |
PREDICTED: uncharacterized protein LOC104088736 isoform X2 [Nicotiana tomentosiformis] |
| 16 |
Hb_005379_010 |
0.2857900139 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_000646_030 |
0.2858532565 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 18 |
Hb_025662_020 |
0.2858572311 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_003797_030 |
0.2858736519 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_000329_460 |
0.2922222241 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Cucumis sativus] |