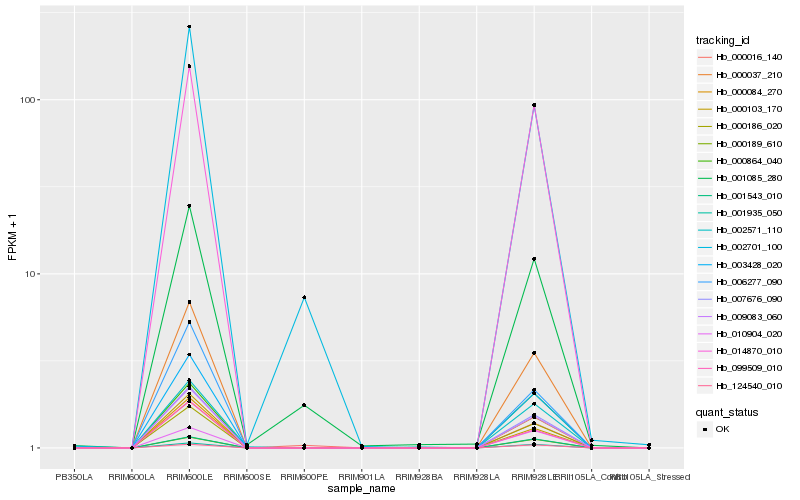
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104901327 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_000084_270 |
0.0071808947 |
- |
- |
hypothetical protein POPTR_0006s22820g [Populus trichocarpa] |
| 3 |
Hb_014870_010 |
0.0297821165 |
- |
- |
PREDICTED: uncharacterized protein LOC104891359 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_007676_090 |
0.0301737828 |
- |
- |
PREDICTED: uncharacterized protein LOC105644038 [Jatropha curcas] |
| 5 |
Hb_003428_020 |
0.034565044 |
- |
- |
PREDICTED: uncharacterized protein LOC105140247 [Populus euphratica] |
| 6 |
Hb_000037_210 |
0.0371720097 |
- |
- |
- |
| 7 |
Hb_009083_060 |
0.0380225125 |
- |
- |
PREDICTED: uncharacterized protein LOC103344557 [Prunus mume] |
| 8 |
Hb_001935_050 |
0.065625197 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 7 [Jatropha curcas] |
| 9 |
Hb_002571_110 |
0.0791821875 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 10 |
Hb_000864_040 |
0.0844951995 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000189_610 |
0.0889655853 |
- |
- |
Late embryogenesis abundant protein D-34, putative [Ricinus communis] |
| 12 |
Hb_001543_010 |
0.0901684794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_099509_010 |
0.0958321178 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_010904_020 |
0.0962725583 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 [Populus euphratica] |
| 15 |
Hb_124540_010 |
0.0986154676 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1-like [Populus euphratica] |
| 16 |
Hb_000103_170 |
0.1139120174 |
- |
- |
uncharacterized protein LOC100501254 [Zea mays] |
| 17 |
Hb_001085_280 |
0.1195176621 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 18 |
Hb_000016_140 |
0.1203314611 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 19 |
Hb_006277_090 |
0.1212034018 |
- |
- |
PREDICTED: uncharacterized protein LOC105649700 [Jatropha curcas] |
| 20 |
Hb_002701_100 |
0.1215288695 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |