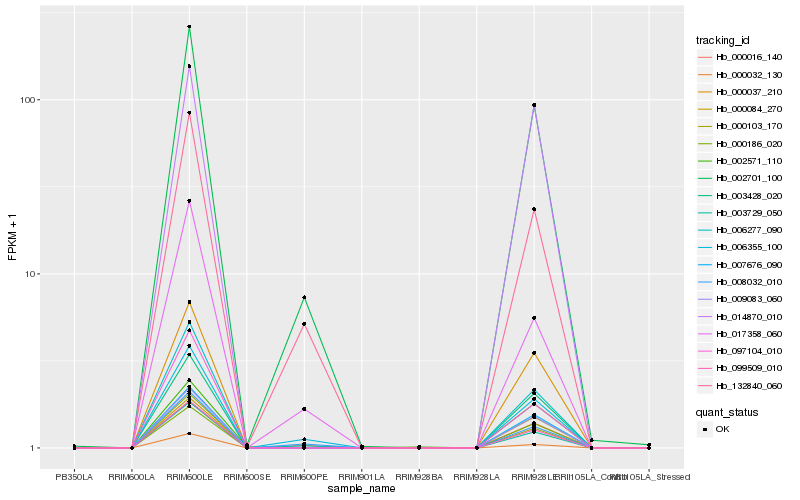
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_099509_010 |
0.0 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000103_170 |
0.0181878069 |
- |
- |
uncharacterized protein LOC100501254 [Zea mays] |
| 3 |
Hb_006277_090 |
0.0255341017 |
- |
- |
PREDICTED: uncharacterized protein LOC105649700 [Jatropha curcas] |
| 4 |
Hb_009083_060 |
0.0579135065 |
- |
- |
PREDICTED: uncharacterized protein LOC103344557 [Prunus mume] |
| 5 |
Hb_000032_130 |
0.0585938837 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 12 [Jatropha curcas] |
| 6 |
Hb_000037_210 |
0.0587630677 |
- |
- |
- |
| 7 |
Hb_003428_020 |
0.0613667355 |
- |
- |
PREDICTED: uncharacterized protein LOC105140247 [Populus euphratica] |
| 8 |
Hb_007676_090 |
0.0657510533 |
- |
- |
PREDICTED: uncharacterized protein LOC105644038 [Jatropha curcas] |
| 9 |
Hb_097104_010 |
0.0663413774 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 10 |
Hb_000186_020 |
0.0958321178 |
- |
- |
PREDICTED: uncharacterized protein LOC104901327 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_002701_100 |
0.0978735039 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 12 |
Hb_000084_270 |
0.1029799415 |
- |
- |
hypothetical protein POPTR_0006s22820g [Populus trichocarpa] |
| 13 |
Hb_003729_050 |
0.1157055727 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_014870_010 |
0.1254513181 |
- |
- |
PREDICTED: uncharacterized protein LOC104891359 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_006355_100 |
0.1254564765 |
- |
- |
hypothetical protein glysoja_002455 [Glycine soja] |
| 16 |
Hb_002571_110 |
0.1333677502 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 17 |
Hb_008032_010 |
0.134772686 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 18 |
Hb_000016_140 |
0.1371238949 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 19 |
Hb_017358_060 |
0.138930708 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 20 |
Hb_132840_060 |
0.1409984559 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |