| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
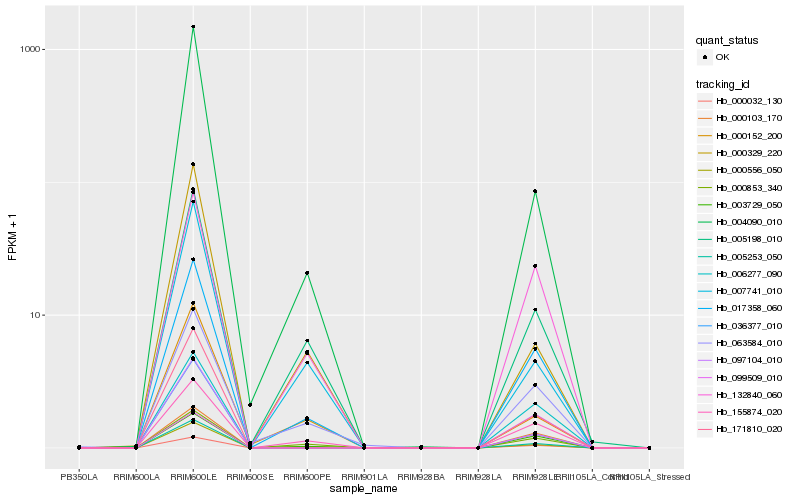
Hb_005253_050 |
0.0 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 2 |
Hb_171810_020 |
0.0160127677 |
- |
- |
peptide/glutathione transporter OPT1 [Vitis vinifera] |
| 3 |
Hb_036377_010 |
0.0642495852 |
- |
- |
PREDICTED: uncharacterized protein LOC105649946 [Jatropha curcas] |
| 4 |
Hb_097104_010 |
0.0771686979 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 5 |
Hb_000032_130 |
0.0848890254 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 12 [Jatropha curcas] |
| 6 |
Hb_017358_060 |
0.1176240726 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 7 |
Hb_006277_090 |
0.1176859273 |
- |
- |
PREDICTED: uncharacterized protein LOC105649700 [Jatropha curcas] |
| 8 |
Hb_004090_010 |
0.120890787 |
- |
- |
Major latex protein, putative [Ricinus communis] |
| 9 |
Hb_000103_170 |
0.1249445428 |
- |
- |
uncharacterized protein LOC100501254 [Zea mays] |
| 10 |
Hb_000556_050 |
0.141562525 |
- |
- |
- |
| 11 |
Hb_099509_010 |
0.1428747321 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_005198_010 |
0.1663996988 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 13 |
Hb_003729_050 |
0.1687606469 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000329_220 |
0.1750088067 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 1 [Jatropha curcas] |
| 15 |
Hb_155874_020 |
0.1756748527 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 16 |
Hb_063584_010 |
0.1785154004 |
- |
- |
hypothetical protein JCGZ_10530 [Jatropha curcas] |
| 17 |
Hb_007741_010 |
0.1801769871 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 1 [Jatropha curcas] |
| 18 |
Hb_132840_060 |
0.1808081337 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 19 |
Hb_000152_200 |
0.1830043188 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase ERL2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000853_340 |
0.1834272162 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X2 [Populus euphratica] |