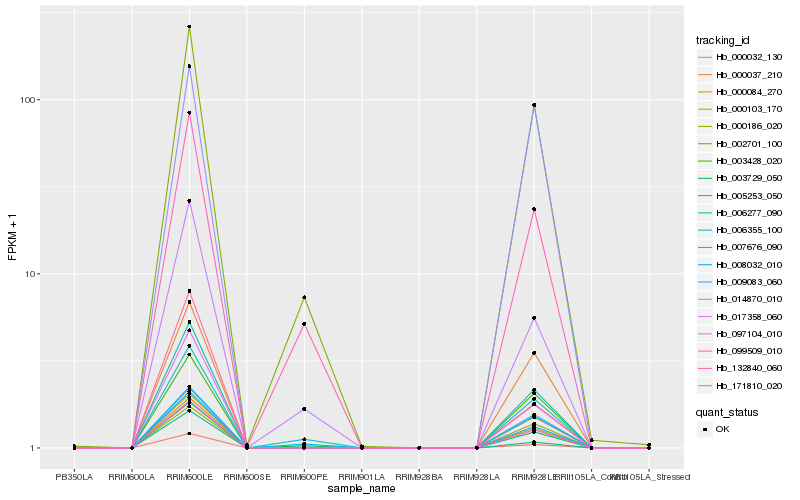
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_170 |
0.0 |
- |
- |
uncharacterized protein LOC100501254 [Zea mays] |
| 2 |
Hb_006277_090 |
0.0073485167 |
- |
- |
PREDICTED: uncharacterized protein LOC105649700 [Jatropha curcas] |
| 3 |
Hb_099509_010 |
0.0181878069 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000032_130 |
0.0404356745 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 12 [Jatropha curcas] |
| 5 |
Hb_097104_010 |
0.0481940258 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 6 |
Hb_009083_060 |
0.0760553747 |
- |
- |
PREDICTED: uncharacterized protein LOC103344557 [Prunus mume] |
| 7 |
Hb_000037_210 |
0.0769037998 |
- |
- |
- |
| 8 |
Hb_003428_020 |
0.0795039107 |
- |
- |
PREDICTED: uncharacterized protein LOC105140247 [Populus euphratica] |
| 9 |
Hb_007676_090 |
0.0838819837 |
- |
- |
PREDICTED: uncharacterized protein LOC105644038 [Jatropha curcas] |
| 10 |
Hb_002701_100 |
0.1032007811 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 11 |
Hb_000186_020 |
0.1139120174 |
- |
- |
PREDICTED: uncharacterized protein LOC104901327 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_003729_050 |
0.1142830787 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000084_270 |
0.1210457349 |
- |
- |
hypothetical protein POPTR_0006s22820g [Populus trichocarpa] |
| 14 |
Hb_005253_050 |
0.1249445428 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 15 |
Hb_006355_100 |
0.1272773198 |
- |
- |
hypothetical protein glysoja_002455 [Glycine soja] |
| 16 |
Hb_017358_060 |
0.1279427354 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 17 |
Hb_008032_010 |
0.1346909671 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 18 |
Hb_132840_060 |
0.1388326594 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 19 |
Hb_171810_020 |
0.1406511193 |
- |
- |
peptide/glutathione transporter OPT1 [Vitis vinifera] |
| 20 |
Hb_014870_010 |
0.1434682017 |
- |
- |
PREDICTED: uncharacterized protein LOC104891359 [Beta vulgaris subsp. vulgaris] |