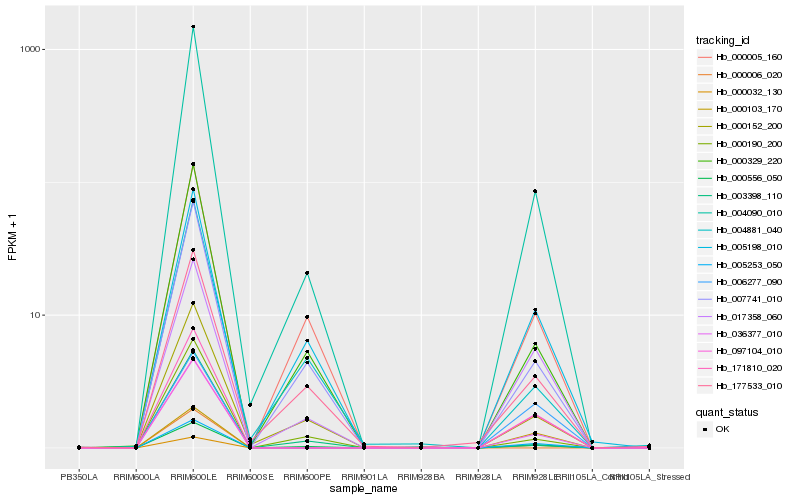
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_036377_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649946 [Jatropha curcas] |
| 2 |
Hb_171810_020 |
0.0483220257 |
- |
- |
peptide/glutathione transporter OPT1 [Vitis vinifera] |
| 3 |
Hb_005253_050 |
0.0642495852 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 4 |
Hb_004090_010 |
0.085371424 |
- |
- |
Major latex protein, putative [Ricinus communis] |
| 5 |
Hb_000329_220 |
0.1379710888 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 1 [Jatropha curcas] |
| 6 |
Hb_097104_010 |
0.1404773121 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 7 |
Hb_000556_050 |
0.1461021211 |
- |
- |
- |
| 8 |
Hb_000032_130 |
0.148067809 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 12 [Jatropha curcas] |
| 9 |
Hb_007741_010 |
0.1539668011 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 1 [Jatropha curcas] |
| 10 |
Hb_017358_060 |
0.1564292991 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 11 |
Hb_000190_200 |
0.1598698217 |
- |
- |
PREDICTED: uncharacterized protein LOC105649946 [Jatropha curcas] |
| 12 |
Hb_000152_200 |
0.1670263612 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase ERL2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000005_160 |
0.1699060405 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 14 |
Hb_005198_010 |
0.1734625854 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 15 |
Hb_006277_090 |
0.1802636477 |
- |
- |
PREDICTED: uncharacterized protein LOC105649700 [Jatropha curcas] |
| 16 |
Hb_177533_010 |
0.1810096748 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 17 |
Hb_003398_110 |
0.1814558985 |
- |
- |
PREDICTED: uncharacterized protein LOC105647583 [Jatropha curcas] |
| 18 |
Hb_004881_040 |
0.1842512092 |
- |
- |
PREDICTED: protodermal factor 1-like [Populus euphratica] |
| 19 |
Hb_000103_170 |
0.1873799594 |
- |
- |
uncharacterized protein LOC100501254 [Zea mays] |
| 20 |
Hb_000006_020 |
0.1874194823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |