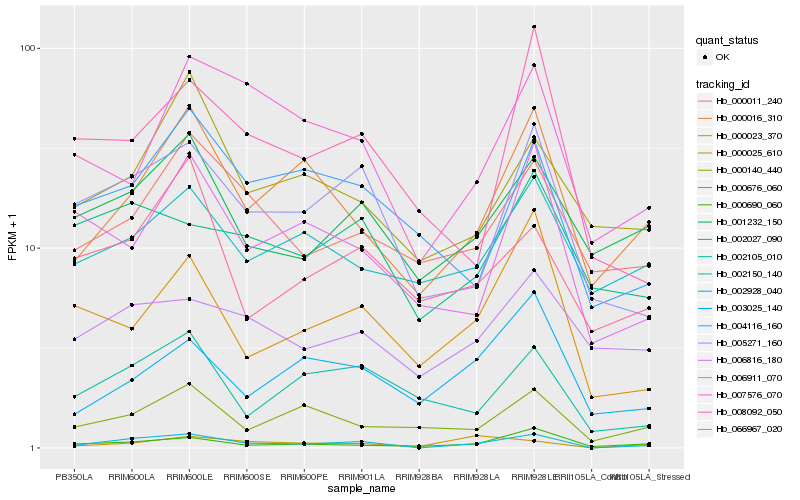
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002928_040 |
0.0 |
- |
- |
PREDICTED: vinorine synthase-like [Jatropha curcas] |
| 2 |
Hb_005271_160 |
0.2131618683 |
- |
- |
PREDICTED: uncharacterized protein LOC105645844 [Jatropha curcas] |
| 3 |
Hb_000016_310 |
0.220210278 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_000690_060 |
0.2389074228 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 5 |
Hb_002105_010 |
0.2444881359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001232_150 |
0.2449323347 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 7 |
Hb_000023_370 |
0.2456125744 |
- |
- |
- |
| 8 |
Hb_000676_060 |
0.2474675633 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 9 |
Hb_006911_070 |
0.2494940836 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 10 |
Hb_007576_070 |
0.2498868671 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000011_240 |
0.2509265095 |
- |
- |
PREDICTED: uncharacterized protein LOC105631316 [Jatropha curcas] |
| 12 |
Hb_004116_160 |
0.2516446575 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000140_440 |
0.2571387904 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 14 |
Hb_003025_140 |
0.257182547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_066967_020 |
0.257633203 |
- |
- |
RAB6-interacting protein, putative [Ricinus communis] |
| 16 |
Hb_008092_050 |
0.2580196206 |
- |
- |
PREDICTED: uncharacterized protein LOC105142434 [Populus euphratica] |
| 17 |
Hb_002027_090 |
0.2629509709 |
- |
- |
- |
| 18 |
Hb_000025_610 |
0.2629677274 |
- |
- |
annexin, putative [Ricinus communis] |
| 19 |
Hb_002150_140 |
0.2649457763 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 20 |
Hb_006816_180 |
0.2650720617 |
- |
- |
- |