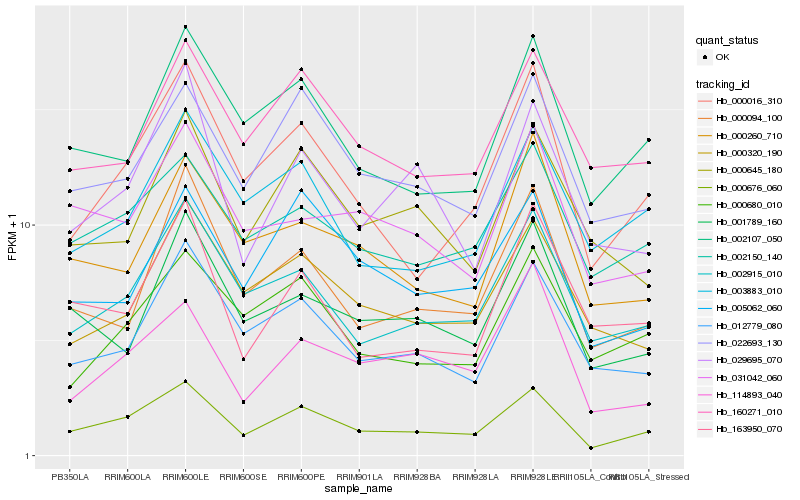
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000676_060 |
0.0 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 2 |
Hb_000016_310 |
0.1023203459 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_002915_010 |
0.1045749649 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 4 |
Hb_022693_130 |
0.1072674889 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_002150_140 |
0.1102596588 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 6 |
Hb_002107_050 |
0.1122456696 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 7 |
Hb_012779_080 |
0.1163790962 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 8 |
Hb_160271_010 |
0.1225388313 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_003883_010 |
0.1245396963 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 10 |
Hb_005062_060 |
0.1255050038 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 11 |
Hb_000094_100 |
0.1263550504 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 12 |
Hb_000320_190 |
0.1274358747 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_029695_070 |
0.1277353573 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000680_010 |
0.1282622698 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Populus euphratica] |
| 15 |
Hb_031042_060 |
0.1299704915 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_001789_160 |
0.132482531 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |
| 17 |
Hb_000645_180 |
0.1342189705 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 18 |
Hb_163950_070 |
0.1347183634 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 19 |
Hb_000260_710 |
0.1349528494 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 20 |
Hb_114893_040 |
0.1352637808 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |