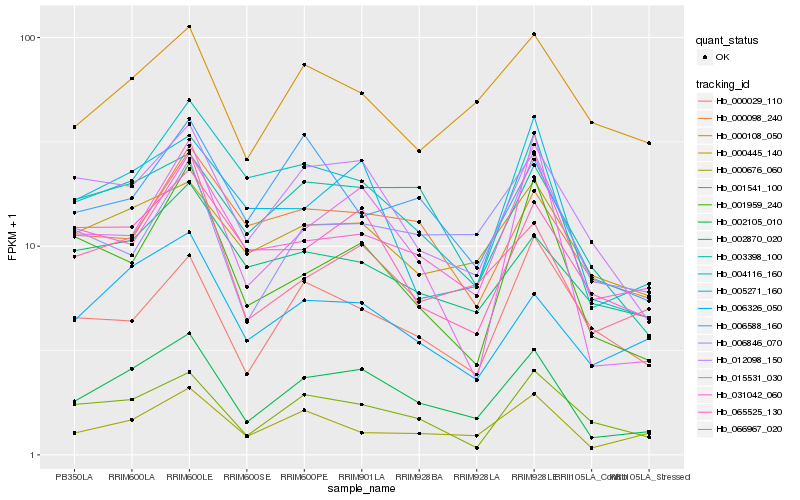
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002105_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012098_150 |
0.1244013261 |
- |
- |
ARC5 protein [Manihot esculenta] |
| 3 |
Hb_004116_160 |
0.1294625342 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000676_060 |
0.1364252875 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 5 |
Hb_066967_020 |
0.1367798955 |
- |
- |
RAB6-interacting protein, putative [Ricinus communis] |
| 6 |
Hb_006326_050 |
0.1398900981 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_005271_160 |
0.1412328638 |
- |
- |
PREDICTED: uncharacterized protein LOC105645844 [Jatropha curcas] |
| 8 |
Hb_015531_030 |
0.1417937235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003398_100 |
0.1434731773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001959_240 |
0.1445924471 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit B, chloroplastic/mitochondrial [Jatropha curcas] |
| 11 |
Hb_001541_100 |
0.1447541594 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 12 |
Hb_000445_140 |
0.1455354398 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 13 |
Hb_031042_060 |
0.1468995757 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 14 |
Hb_065525_130 |
0.1499668142 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 15 |
Hb_006588_160 |
0.1563927912 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_002870_020 |
0.1572271999 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000108_050 |
0.1584198818 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 18 |
Hb_000098_240 |
0.1584885996 |
- |
- |
PREDICTED: UDP-glucose:glycoprotein glucosyltransferase [Jatropha curcas] |
| 19 |
Hb_000029_110 |
0.1611059893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006846_070 |
0.1618345869 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |