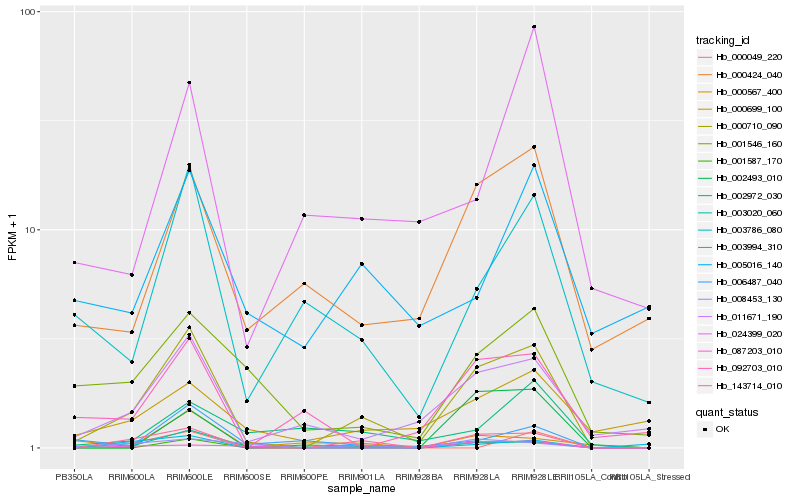
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000710_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104583249 [Brachypodium distachyon] |
| 2 |
Hb_143714_010 |
0.178187987 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Populus euphratica] |
| 3 |
Hb_003020_060 |
0.2644428915 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein [Jatropha curcas] |
| 4 |
Hb_011671_190 |
0.2680489351 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 5 |
Hb_008453_130 |
0.2944370888 |
- |
- |
hypothetical protein JCGZ_08179 [Jatropha curcas] |
| 6 |
Hb_003786_080 |
0.2990743387 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 7 |
Hb_000424_040 |
0.3072170259 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 8 |
Hb_001546_160 |
0.3081400081 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_001587_170 |
0.3230438411 |
- |
- |
hypothetical protein B456_010G223200, partial [Gossypium raimondii] |
| 10 |
Hb_092703_010 |
0.3263091794 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 11 |
Hb_000699_100 |
0.3266727926 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01510, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_006487_040 |
0.3271055595 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 13 |
Hb_002972_030 |
0.3357497056 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_002493_010 |
0.3378201134 |
- |
- |
- |
| 15 |
Hb_003994_310 |
0.3384729984 |
transcription factor |
TF Family: PHD |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_024399_020 |
0.33973625 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 17 |
Hb_087203_010 |
0.3432270271 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 68-like [Pyrus x bretschneideri] |
| 18 |
Hb_000567_400 |
0.3512246421 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 19 |
Hb_000049_220 |
0.3522307627 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 20 |
Hb_005016_140 |
0.3555702612 |
- |
- |
PREDICTED: thylakoid lumenal 15.0 kDa protein 2, chloroplastic isoform X1 [Jatropha curcas] |