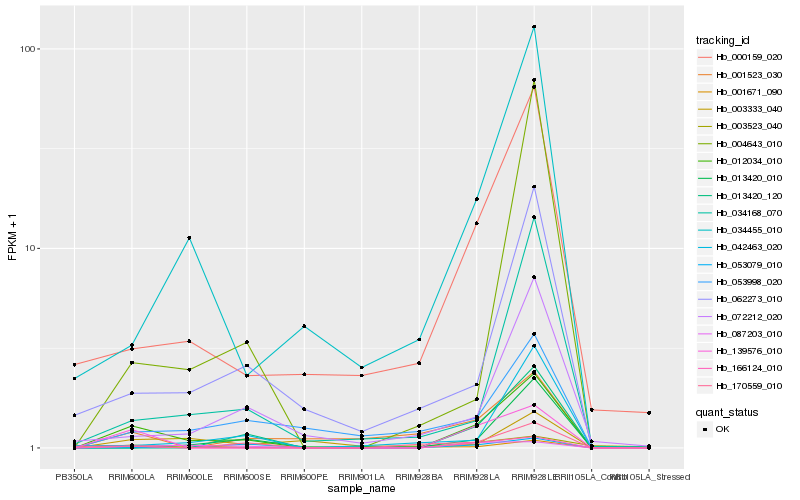
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012034_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_139576_010 |
0.2455181932 |
- |
- |
PREDICTED: uncharacterized protein LOC105795953 [Gossypium raimondii] |
| 3 |
Hb_166124_010 |
0.2520747288 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 4 |
Hb_000159_020 |
0.2802980055 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Jatropha curcas] |
| 5 |
Hb_062273_010 |
0.2910762521 |
- |
- |
ATP synthase F0 subunit 9 [Chara vulgaris] |
| 6 |
Hb_087203_010 |
0.2911808695 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 68-like [Pyrus x bretschneideri] |
| 7 |
Hb_034455_010 |
0.2930957994 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_001671_090 |
0.2961466839 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_034168_070 |
0.3025813665 |
- |
- |
ATP synthase F1 subunit 1 (mitochondrion) [Hevea brasiliensis] |
| 10 |
Hb_053998_020 |
0.3065180185 |
- |
- |
- |
| 11 |
Hb_072212_020 |
0.3092627664 |
- |
- |
hypothetical protein B456_001G162300 [Gossypium raimondii] |
| 12 |
Hb_013420_120 |
0.311616259 |
- |
- |
- |
| 13 |
Hb_003523_040 |
0.3154359075 |
- |
- |
F15O4.13 [Arabidopsis thaliana] |
| 14 |
Hb_053079_010 |
0.3171775373 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 15 |
Hb_013420_010 |
0.3185767646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_042463_020 |
0.3243463467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_170559_010 |
0.3254276851 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 18 |
Hb_001523_030 |
0.3265984203 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_004643_010 |
0.3267750503 |
- |
- |
cytochrome B, putative [Ricinus communis] |
| 20 |
Hb_003333_040 |
0.3302865512 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |