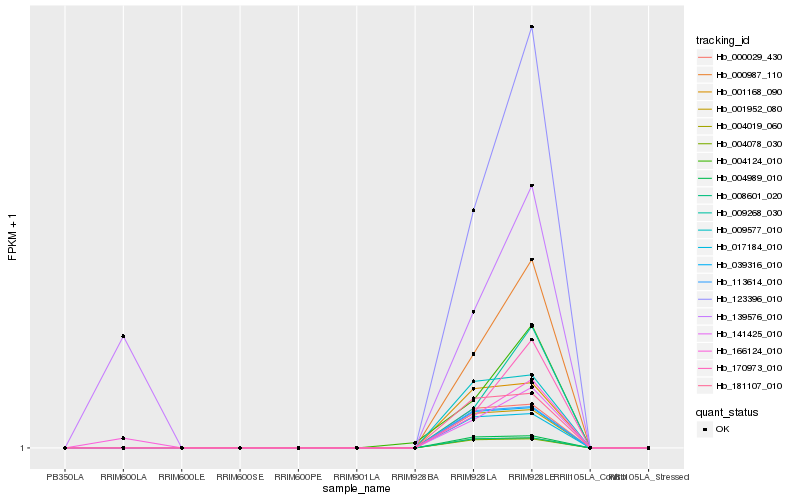
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_166124_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 2 |
Hb_139576_010 |
0.1421708221 |
- |
- |
PREDICTED: uncharacterized protein LOC105795953 [Gossypium raimondii] |
| 3 |
Hb_141425_010 |
0.209488112 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |
| 4 |
Hb_000987_110 |
0.209495239 |
- |
- |
PREDICTED: uncharacterized protein LOC104902780 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_123396_010 |
0.2095902804 |
- |
- |
hypothetical protein AMTR_s00510p00011440 [Amborella trichopoda] |
| 6 |
Hb_009268_030 |
0.2222777628 |
- |
- |
ribosomal protein S7 [Piper cenocladum] |
| 7 |
Hb_170973_010 |
0.2223157393 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |
| 8 |
Hb_004124_010 |
0.2420205416 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 9 |
Hb_004078_030 |
0.2511503015 |
- |
- |
Putative polyprotein, identical [Solanum demissum] |
| 10 |
Hb_004019_060 |
0.2511617929 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 11 |
Hb_008601_020 |
0.2511650707 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_004989_010 |
0.2511924344 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy sub-class [Oryza sativa Japonica Group] |
| 13 |
Hb_017184_010 |
0.2514995897 |
- |
- |
PREDICTED: uncharacterized protein LOC105778857 [Gossypium raimondii] |
| 14 |
Hb_001952_080 |
0.2515561156 |
- |
- |
PREDICTED: uncharacterized protein LOC105042236 isoform X2 [Elaeis guineensis] |
| 15 |
Hb_039316_010 |
0.2515840495 |
- |
- |
PREDICTED: uncharacterized protein LOC105771917 [Gossypium raimondii] |
| 16 |
Hb_113614_010 |
0.2515996739 |
- |
- |
hypothetical protein VITISV_001308 [Vitis vinifera] |
| 17 |
Hb_000029_430 |
0.2516347357 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_181107_010 |
0.2517944908 |
- |
- |
hypothetical protein, partial [Pseudomonas tolaasii] |
| 19 |
Hb_001168_090 |
0.2519525807 |
- |
- |
PREDICTED: uncharacterized protein LOC104246050 [Nicotiana sylvestris] |
| 20 |
Hb_009577_010 |
0.252072865 |
- |
- |
- |