| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
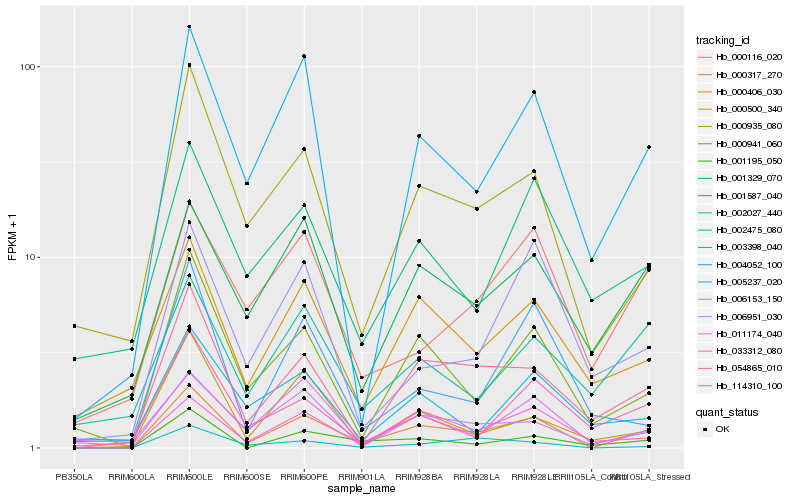
Hb_000317_270 |
0.0 |
- |
- |
Uncharacterized protein TCM_001673 [Theobroma cacao] |
| 2 |
Hb_114310_100 |
0.1719240368 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 3 |
Hb_004052_100 |
0.179663649 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 4 |
Hb_000941_060 |
0.1820763961 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0016s05890g [Populus trichocarpa] |
| 5 |
Hb_033312_080 |
0.1911832199 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 6 |
Hb_000935_080 |
0.1918398944 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_001195_050 |
0.1980761248 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 8 |
Hb_011174_040 |
0.2082488656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001329_070 |
0.2082725752 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 10 |
Hb_000406_030 |
0.212227471 |
- |
- |
Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase [Morus notabilis] |
| 11 |
Hb_002475_080 |
0.2166896583 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein MALE MEIOCYTE DEATH 1 [Jatropha curcas] |
| 12 |
Hb_054865_010 |
0.2197337138 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 13 |
Hb_006951_030 |
0.2217113413 |
- |
- |
PREDICTED: protein MCM10 homolog [Jatropha curcas] |
| 14 |
Hb_000500_340 |
0.2237127132 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 15 |
Hb_006153_150 |
0.2305353286 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 16 |
Hb_003398_040 |
0.2313996021 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001587_040 |
0.2338012505 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000116_020 |
0.2356239545 |
- |
- |
PREDICTED: serine acetyltransferase 2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002027_440 |
0.2363923892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005237_020 |
0.2384711717 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |