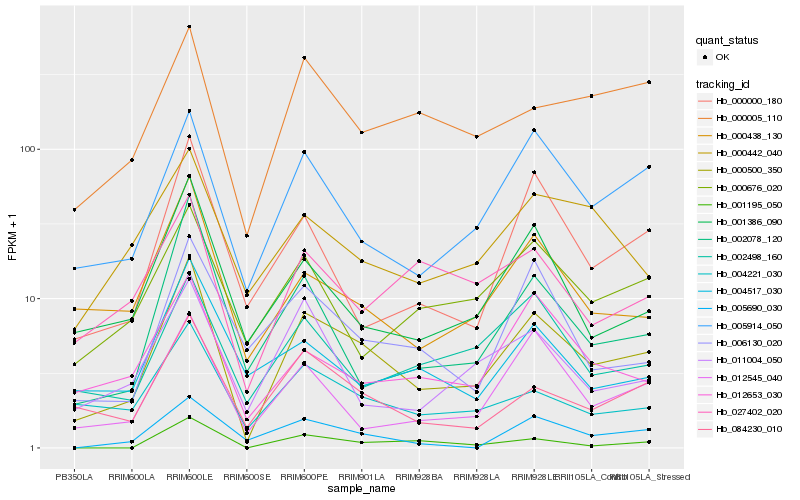
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000500_350 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 2 |
Hb_084230_010 |
0.1662937221 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 3 |
Hb_005914_050 |
0.1699975729 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000438_130 |
0.1756669472 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 5 |
Hb_001386_090 |
0.1778326099 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 6 |
Hb_004221_030 |
0.1800087642 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_012545_040 |
0.1854378876 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002078_120 |
0.1873085766 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 9 |
Hb_000000_180 |
0.1911960192 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 10 |
Hb_000005_110 |
0.1925467726 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 11 |
Hb_001195_050 |
0.1925533403 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 12 |
Hb_012653_030 |
0.1927390705 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 4, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_002498_160 |
0.1944219219 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 14 |
Hb_006130_020 |
0.1968363587 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000676_020 |
0.1993479083 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 16 |
Hb_011004_050 |
0.1998095203 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 17 |
Hb_005690_030 |
0.2016965901 |
- |
- |
hypothetical protein VITISV_020378 [Vitis vinifera] |
| 18 |
Hb_027402_020 |
0.2023847454 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 19 |
Hb_004517_030 |
0.2044219717 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000442_040 |
0.2048680175 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |