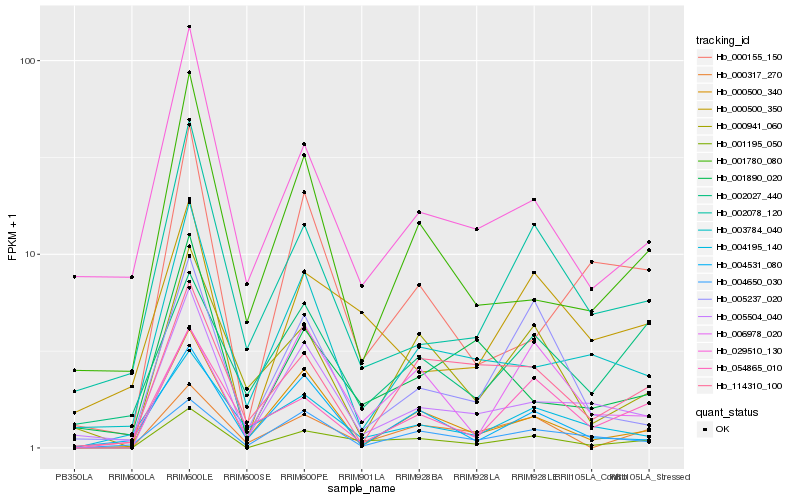
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_050 |
0.0 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 2 |
Hb_000500_350 |
0.1925533403 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 3 |
Hb_000317_270 |
0.1980761248 |
- |
- |
Uncharacterized protein TCM_001673 [Theobroma cacao] |
| 4 |
Hb_000500_340 |
0.2209680906 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 5 |
Hb_005504_040 |
0.22136106 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000155_150 |
0.2302899038 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 7 |
Hb_004650_030 |
0.2314060672 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002078_120 |
0.2337874839 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 9 |
Hb_054865_010 |
0.2375487688 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 10 |
Hb_000941_060 |
0.2400655768 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0016s05890g [Populus trichocarpa] |
| 11 |
Hb_001780_080 |
0.2412272528 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 12 |
Hb_114310_100 |
0.2414575509 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 13 |
Hb_005237_020 |
0.2423618402 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 14 |
Hb_029510_130 |
0.2437074696 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 15 |
Hb_006978_020 |
0.2444664096 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_001890_020 |
0.2455546725 |
- |
- |
hypothetical protein CICLE_v10025198mg [Citrus clementina] |
| 17 |
Hb_003784_040 |
0.2479066895 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 18 |
Hb_004195_140 |
0.2502285546 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 19 |
Hb_002027_440 |
0.2518057027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004531_080 |
0.2519185852 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |