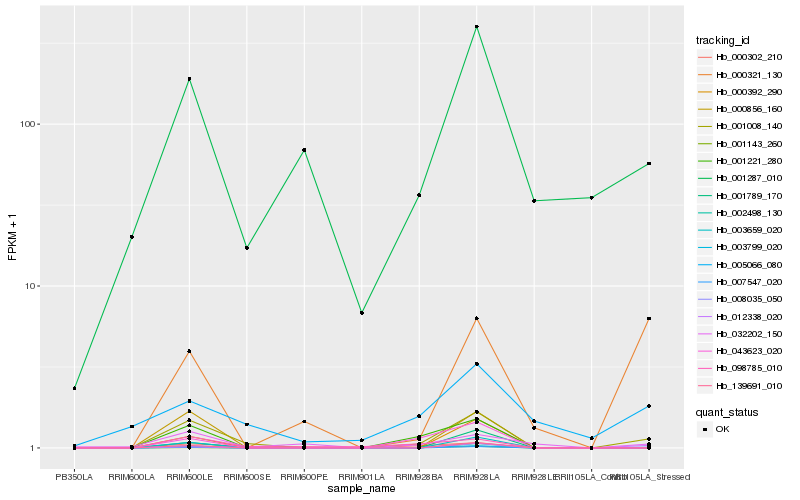
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001008_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_003799_020 |
0.2624529367 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 3 |
Hb_007547_020 |
0.2877627019 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase A-like [Populus euphratica] |
| 4 |
Hb_001221_280 |
0.3168883777 |
- |
- |
- |
| 5 |
Hb_000856_160 |
0.3212506142 |
- |
- |
- |
| 6 |
Hb_003659_020 |
0.3225168353 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 7 |
Hb_008035_050 |
0.3236644922 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 8 |
Hb_139691_010 |
0.3260820994 |
- |
- |
- |
| 9 |
Hb_001287_010 |
0.3313086472 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 10 |
Hb_000392_290 |
0.3418227819 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_012338_020 |
0.3468151197 |
- |
- |
- |
| 12 |
Hb_098785_010 |
0.3505094659 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 13 |
Hb_032202_150 |
0.3528625061 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR14 [Jatropha curcas] |
| 14 |
Hb_001789_170 |
0.3600670509 |
- |
- |
hypothetical protein JCGZ_08269 [Jatropha curcas] |
| 15 |
Hb_000302_210 |
0.3613735224 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_043623_020 |
0.3643342514 |
- |
- |
hypothetical protein JCGZ_17692 [Jatropha curcas] |
| 17 |
Hb_001143_260 |
0.3645126907 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 18 |
Hb_000321_130 |
0.3685362633 |
- |
- |
Alcohol dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_005066_080 |
0.3712733398 |
- |
- |
hypothetical protein PRUPE_ppa025081mg [Prunus persica] |
| 20 |
Hb_002498_130 |
0.3769098392 |
- |
- |
hypothetical protein POPTR_0004s00980g [Populus trichocarpa] |