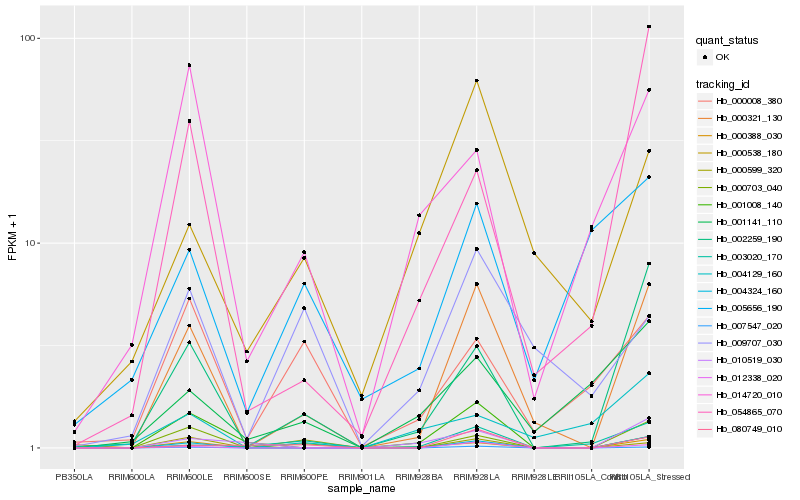
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000321_130 |
0.0 |
- |
- |
Alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_004324_160 |
0.2018596453 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 3 |
Hb_007547_020 |
0.2147023371 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase A-like [Populus euphratica] |
| 4 |
Hb_000703_040 |
0.2292004909 |
- |
- |
hypothetical protein RCOM_1177160 [Ricinus communis] |
| 5 |
Hb_012338_020 |
0.2321397718 |
- |
- |
- |
| 6 |
Hb_002259_190 |
0.244634595 |
- |
- |
Omega-3 fatty acid desaturase, endoplasmic reticulum, putative [Ricinus communis] |
| 7 |
Hb_054865_070 |
0.3079509462 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 8 |
Hb_080749_010 |
0.3106226528 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Populus euphratica] |
| 9 |
Hb_000538_180 |
0.3147119593 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 10 |
Hb_010519_030 |
0.3152731582 |
- |
- |
- |
| 11 |
Hb_009707_030 |
0.3366776903 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 12 |
Hb_003020_170 |
0.3373952252 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 13 |
Hb_001141_110 |
0.3374406279 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 14 |
Hb_000388_030 |
0.3448494604 |
- |
- |
hypothetical protein JCGZ_13427 [Jatropha curcas] |
| 15 |
Hb_014720_010 |
0.3502484401 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004129_160 |
0.3519562796 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 17 |
Hb_000008_380 |
0.3571781032 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000599_320 |
0.3581687523 |
- |
- |
hypothetical protein JCGZ_17662 [Jatropha curcas] |
| 19 |
Hb_005656_190 |
0.3657893943 |
- |
- |
proteasome subunit beta type 6,9, putative [Ricinus communis] |
| 20 |
Hb_001008_140 |
0.3685362633 |
- |
- |
- |