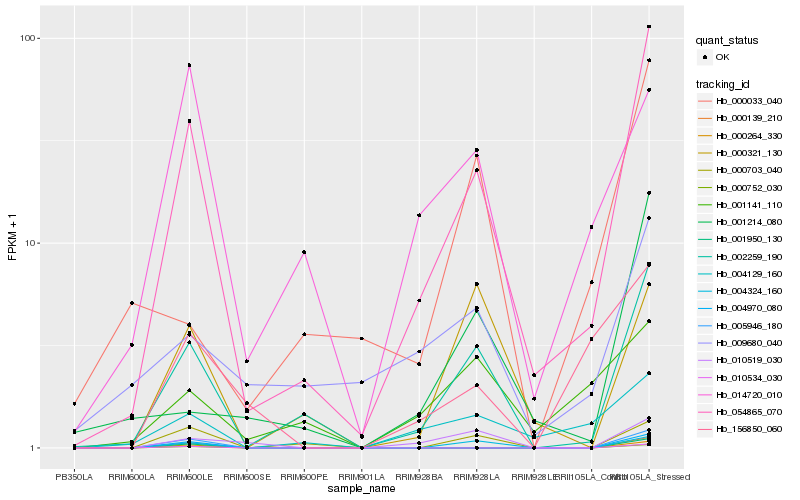
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054865_070 |
0.0 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 2 |
Hb_002259_190 |
0.1450775058 |
- |
- |
Omega-3 fatty acid desaturase, endoplasmic reticulum, putative [Ricinus communis] |
| 3 |
Hb_004324_160 |
0.2516178597 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 4 |
Hb_004129_160 |
0.2651659393 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 5 |
Hb_000703_040 |
0.2658960031 |
- |
- |
hypothetical protein RCOM_1177160 [Ricinus communis] |
| 6 |
Hb_010519_030 |
0.2933527332 |
- |
- |
- |
| 7 |
Hb_001214_080 |
0.2951070643 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38)-like [Jatropha curcas] |
| 8 |
Hb_156850_060 |
0.2988044657 |
- |
- |
PREDICTED: uncharacterized protein LOC105635248 [Jatropha curcas] |
| 9 |
Hb_014720_010 |
0.3070874783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000321_130 |
0.3079509462 |
- |
- |
Alcohol dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_009680_040 |
0.3079747531 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_000752_030 |
0.3091768792 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_0978110 [Ricinus communis] |
| 13 |
Hb_010534_030 |
0.3092229019 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 14 |
Hb_001950_130 |
0.3093646933 |
- |
- |
- |
| 15 |
Hb_004970_080 |
0.3096450388 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 16 |
Hb_000264_330 |
0.3108405018 |
- |
- |
PREDICTED: uncharacterized protein LOC105766768 [Gossypium raimondii] |
| 17 |
Hb_000139_210 |
0.3112370422 |
- |
- |
- |
| 18 |
Hb_005946_180 |
0.3126672978 |
- |
- |
PREDICTED: uncharacterized protein LOC100818788 [Glycine max] |
| 19 |
Hb_001141_110 |
0.3185044104 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 20 |
Hb_000033_040 |
0.3221539595 |
- |
- |
conserved hypothetical protein [Ricinus communis] |