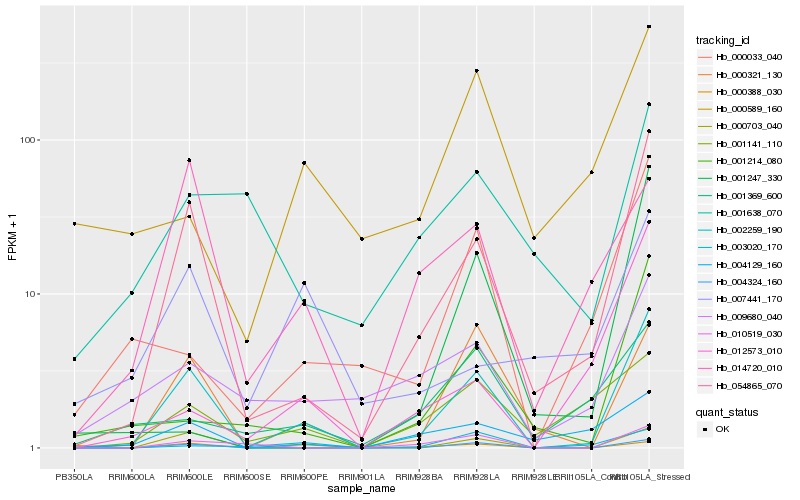
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_190 |
0.0 |
- |
- |
Omega-3 fatty acid desaturase, endoplasmic reticulum, putative [Ricinus communis] |
| 2 |
Hb_054865_070 |
0.1450775058 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 3 |
Hb_000703_040 |
0.1914154075 |
- |
- |
hypothetical protein RCOM_1177160 [Ricinus communis] |
| 4 |
Hb_004324_160 |
0.2178980946 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 5 |
Hb_000321_130 |
0.244634595 |
- |
- |
Alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_010519_030 |
0.2859124529 |
- |
- |
- |
| 7 |
Hb_001214_080 |
0.2946925444 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38)-like [Jatropha curcas] |
| 8 |
Hb_000033_040 |
0.2971065853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004129_160 |
0.2976170324 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 10 |
Hb_009680_040 |
0.3022698238 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 11 |
Hb_014720_010 |
0.3083156548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001141_110 |
0.3127951851 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 13 |
Hb_003020_170 |
0.3273266645 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 14 |
Hb_000589_160 |
0.3298201385 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 15 |
Hb_001369_600 |
0.3340313391 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_001638_070 |
0.3392340898 |
transcription factor |
TF Family: bHLH |
Basic helix-loop-helix DNA-binding family protein [Theobroma cacao] |
| 17 |
Hb_000388_030 |
0.3393353958 |
- |
- |
hypothetical protein JCGZ_13427 [Jatropha curcas] |
| 18 |
Hb_001247_330 |
0.3454241805 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 19 |
Hb_012573_010 |
0.3459028657 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 20 |
Hb_007441_170 |
0.3465182759 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |