| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
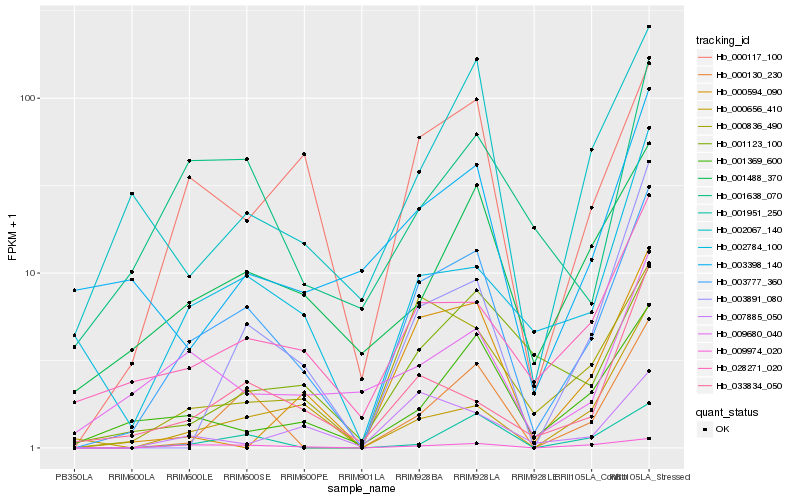
Hb_003777_360 |
0.0 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 2 |
Hb_000117_100 |
0.1837482121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_009974_020 |
0.1873396156 |
- |
- |
PREDICTED: uncharacterized protein LOC104427753 [Eucalyptus grandis] |
| 4 |
Hb_000836_490 |
0.1967879422 |
- |
- |
PREDICTED: E3 SUMO-protein ligase pli1-like [Jatropha curcas] |
| 5 |
Hb_028271_020 |
0.2036967649 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 6 |
Hb_000130_230 |
0.205348851 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098-like [Jatropha curcas] |
| 7 |
Hb_001488_370 |
0.21504386 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 8 |
Hb_003891_080 |
0.2170187889 |
- |
- |
hypothetical protein JCGZ_16710 [Jatropha curcas] |
| 9 |
Hb_002784_100 |
0.2218796744 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 10 |
Hb_001369_600 |
0.2242815978 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_033834_050 |
0.2293570492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002067_140 |
0.2320727417 |
- |
- |
- |
| 13 |
Hb_001951_250 |
0.2411038487 |
- |
- |
- |
| 14 |
Hb_009680_040 |
0.2411827338 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 15 |
Hb_001638_070 |
0.2443073139 |
transcription factor |
TF Family: bHLH |
Basic helix-loop-helix DNA-binding family protein [Theobroma cacao] |
| 16 |
Hb_000594_090 |
0.2464509876 |
- |
- |
PREDICTED: allene oxide cyclase 3, chloroplastic-like [Populus euphratica] |
| 17 |
Hb_007885_050 |
0.2469402619 |
- |
- |
PREDICTED: lignin-forming anionic peroxidase-like [Jatropha curcas] |
| 18 |
Hb_003398_140 |
0.2482074028 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000656_410 |
0.2513169171 |
- |
- |
hypothetical protein POPTR_0003s06410g [Populus trichocarpa] |
| 20 |
Hb_001123_100 |
0.2515350609 |
- |
- |
PREDICTED: uncharacterized protein LOC105647337 isoform X1 [Jatropha curcas] |