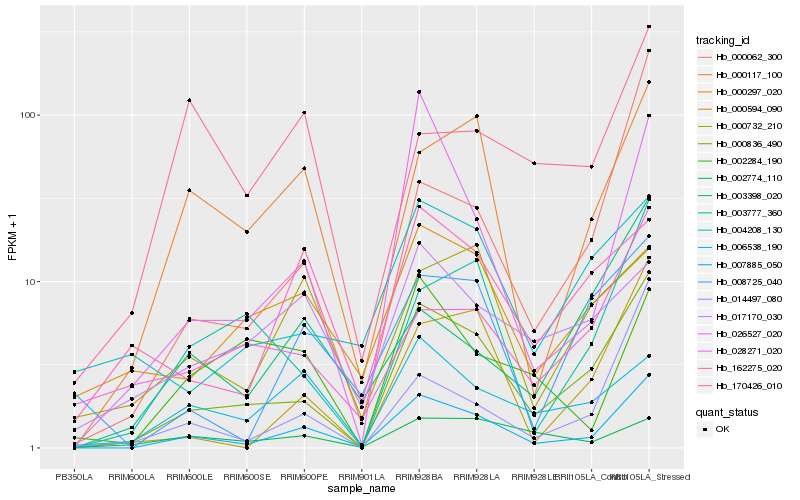
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007885_050 |
0.0 |
- |
- |
PREDICTED: lignin-forming anionic peroxidase-like [Jatropha curcas] |
| 2 |
Hb_000836_490 |
0.1429611715 |
- |
- |
PREDICTED: E3 SUMO-protein ligase pli1-like [Jatropha curcas] |
| 3 |
Hb_000117_100 |
0.2054410335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000594_090 |
0.2131747693 |
- |
- |
PREDICTED: allene oxide cyclase 3, chloroplastic-like [Populus euphratica] |
| 5 |
Hb_026527_020 |
0.2203623044 |
- |
- |
PREDICTED: putative methyltransferase DDB_G0268948 [Jatropha curcas] |
| 6 |
Hb_008725_040 |
0.2322958804 |
- |
- |
PREDICTED: uncharacterized protein At4g29660 [Nelumbo nucifera] |
| 7 |
Hb_006538_190 |
0.2427069442 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003777_360 |
0.2469402619 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 9 |
Hb_014497_080 |
0.2474427581 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 10 |
Hb_162275_020 |
0.2537213877 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 11 |
Hb_000732_210 |
0.2590414692 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |
| 12 |
Hb_000062_300 |
0.2605036466 |
- |
- |
PREDICTED: uncharacterized protein LOC105644761 [Jatropha curcas] |
| 13 |
Hb_004208_130 |
0.2624303821 |
- |
- |
hypothetical protein JCGZ_16532 [Jatropha curcas] |
| 14 |
Hb_028271_020 |
0.2640093373 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 15 |
Hb_017170_030 |
0.2661101655 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 16 |
Hb_003398_020 |
0.26644029 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 17 |
Hb_002284_190 |
0.2751118619 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 18 |
Hb_002774_110 |
0.2765209099 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_000297_020 |
0.2798735571 |
- |
- |
PREDICTED: protein phosphatase 2C 77 [Jatropha curcas] |
| 20 |
Hb_170426_010 |
0.2800327319 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |