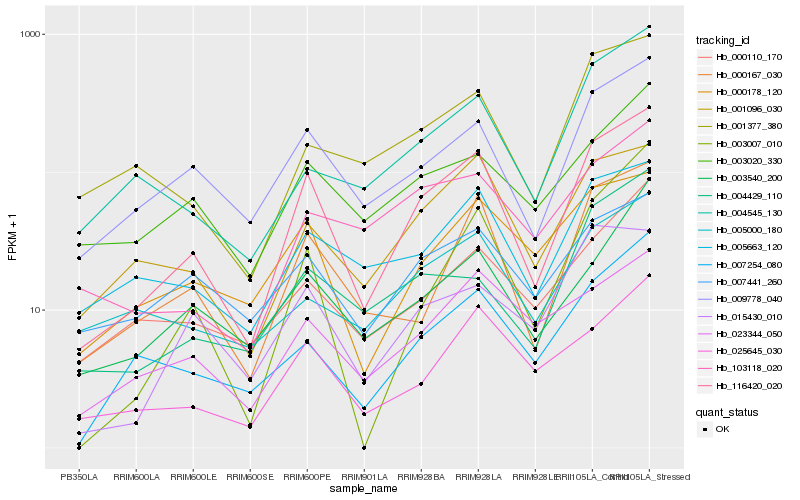
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116420_020 |
0.0 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 2 |
Hb_009778_040 |
0.134713903 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 3 |
Hb_000167_030 |
0.1375438668 |
- |
- |
PREDICTED: uncharacterized protein LOC105641111 isoform X1 [Jatropha curcas] |
| 4 |
Hb_025645_030 |
0.1478599911 |
- |
- |
- |
| 5 |
Hb_001096_030 |
0.1489897922 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |
| 6 |
Hb_007254_080 |
0.1581127452 |
- |
- |
- |
| 7 |
Hb_005663_120 |
0.1613005775 |
- |
- |
PREDICTED: uncharacterized protein LOC105635265 [Jatropha curcas] |
| 8 |
Hb_000178_120 |
0.1622410955 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 9 |
Hb_023344_050 |
0.168531953 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 10 |
Hb_003020_330 |
0.1708115865 |
- |
- |
hypothetical protein B456_010G173100 [Gossypium raimondii] |
| 11 |
Hb_000110_170 |
0.1712441355 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_003540_200 |
0.1719940428 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 13 |
Hb_001377_380 |
0.1721969066 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |
| 14 |
Hb_003007_010 |
0.1724408361 |
- |
- |
PREDICTED: probable peptide/nitrate transporter At3g43790 isoform X1 [Populus euphratica] |
| 15 |
Hb_004545_130 |
0.1734246968 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 16 |
Hb_103118_020 |
0.1751346742 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 17 |
Hb_015430_010 |
0.1752871051 |
- |
- |
PREDICTED: uncharacterized protein LOC105125506 isoform X2 [Populus euphratica] |
| 18 |
Hb_007441_260 |
0.1785152705 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_005000_180 |
0.1788008122 |
- |
- |
hypothetical protein JCGZ_07408 [Jatropha curcas] |
| 20 |
Hb_004429_110 |
0.1796897956 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |