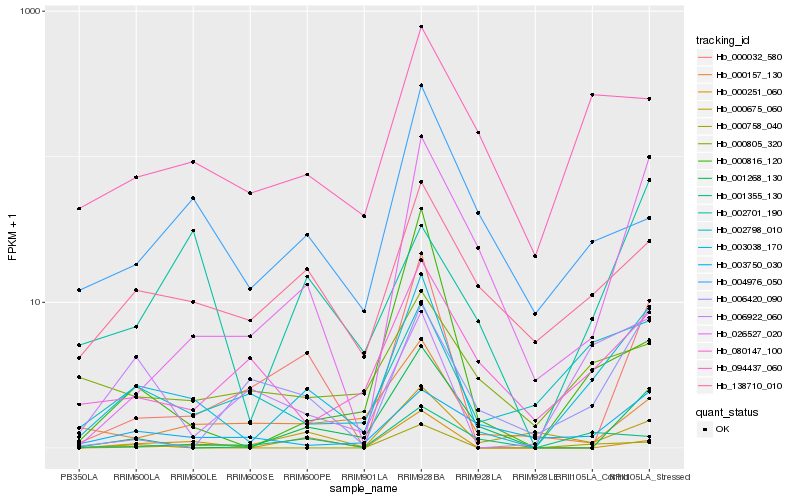
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003038_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_026527_020 |
0.2674214602 |
- |
- |
PREDICTED: putative methyltransferase DDB_G0268948 [Jatropha curcas] |
| 3 |
Hb_138710_010 |
0.2674271217 |
- |
- |
Adenosylmethionine-8-amino-7-oxononanoate aminotransferase [Gossypium arboreum] |
| 4 |
Hb_002798_010 |
0.270624893 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 5 |
Hb_094437_060 |
0.2742338332 |
- |
- |
PREDICTED: cytochrome B5 isoform D-like [Jatropha curcas] |
| 6 |
Hb_000032_580 |
0.2763235247 |
- |
- |
hypothetical protein RCOM_1311780 [Ricinus communis] |
| 7 |
Hb_001268_130 |
0.2834404792 |
- |
- |
hypothetical protein JCGZ_26837 [Jatropha curcas] |
| 8 |
Hb_006922_060 |
0.288218668 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 9 |
Hb_001355_130 |
0.2901742722 |
- |
- |
PREDICTED: uncharacterized protein LOC105137896 [Populus euphratica] |
| 10 |
Hb_003750_030 |
0.2979536744 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 11 |
Hb_080147_100 |
0.2990744311 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN [Jatropha curcas] |
| 12 |
Hb_000758_040 |
0.2995401942 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098 [Jatropha curcas] |
| 13 |
Hb_000816_120 |
0.3070686836 |
- |
- |
PREDICTED: uncharacterized protein LOC105649214 [Jatropha curcas] |
| 14 |
Hb_006420_090 |
0.3091648749 |
- |
- |
PREDICTED: zinc finger protein 5 [Jatropha curcas] |
| 15 |
Hb_004976_050 |
0.3143002056 |
- |
- |
- |
| 16 |
Hb_000805_320 |
0.3156195586 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 17 |
Hb_000675_060 |
0.3170772458 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_000251_060 |
0.3256138307 |
- |
- |
PREDICTED: uncharacterized protein LOC105649757 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000157_130 |
0.3264577437 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X2 [Populus euphratica] |
| 20 |
Hb_002701_190 |
0.3277890752 |
- |
- |
PREDICTED: uncharacterized protein LOC105646742 [Jatropha curcas] |