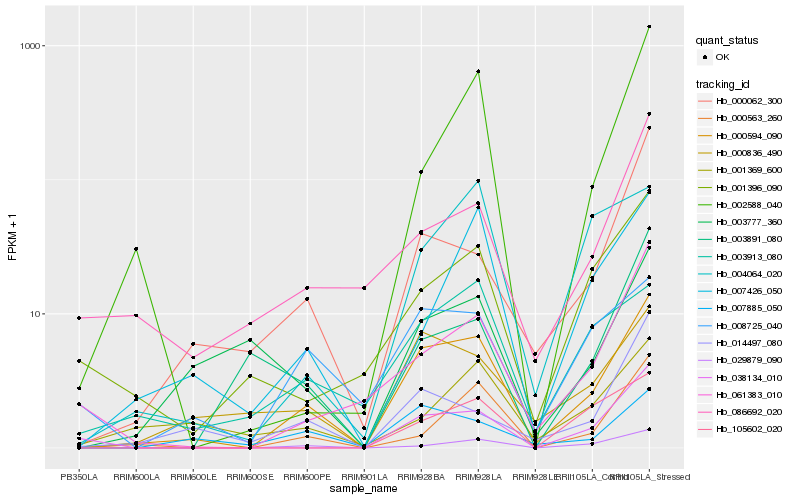
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000594_090 |
0.0 |
- |
- |
PREDICTED: allene oxide cyclase 3, chloroplastic-like [Populus euphratica] |
| 2 |
Hb_029879_090 |
0.1972481195 |
- |
- |
PREDICTED: serine/threonine-protein kinase At3g07070 [Jatropha curcas] |
| 3 |
Hb_000836_490 |
0.2031361721 |
- |
- |
PREDICTED: E3 SUMO-protein ligase pli1-like [Jatropha curcas] |
| 4 |
Hb_007885_050 |
0.2131747693 |
- |
- |
PREDICTED: lignin-forming anionic peroxidase-like [Jatropha curcas] |
| 5 |
Hb_038134_010 |
0.2258430307 |
- |
- |
- |
| 6 |
Hb_000563_260 |
0.2261971418 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 7 |
Hb_008725_040 |
0.2309165519 |
- |
- |
PREDICTED: uncharacterized protein At4g29660 [Nelumbo nucifera] |
| 8 |
Hb_000062_300 |
0.2339842391 |
- |
- |
PREDICTED: uncharacterized protein LOC105644761 [Jatropha curcas] |
| 9 |
Hb_061383_010 |
0.234177991 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 10 |
Hb_007426_050 |
0.2397528076 |
transcription factor |
TF Family: Tify |
JAZ8 [Hevea brasiliensis] |
| 11 |
Hb_001396_090 |
0.240967017 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Jatropha curcas] |
| 12 |
Hb_105602_020 |
0.2416914491 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1-like [Populus euphratica] |
| 13 |
Hb_003913_080 |
0.2418010853 |
- |
- |
rpl27A [Hemiselmis andersenii] |
| 14 |
Hb_003777_360 |
0.2464509876 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 15 |
Hb_014497_080 |
0.2488412132 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 16 |
Hb_002588_040 |
0.2490758385 |
- |
- |
Pathogenesis-related protein 1 [Theobroma cacao] |
| 17 |
Hb_086692_020 |
0.2491985677 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_001369_600 |
0.2581739872 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_004064_020 |
0.2586293067 |
- |
- |
hypothetical protein M569_06853, partial [Genlisea aurea] |
| 20 |
Hb_003891_080 |
0.2595746305 |
- |
- |
hypothetical protein JCGZ_16710 [Jatropha curcas] |