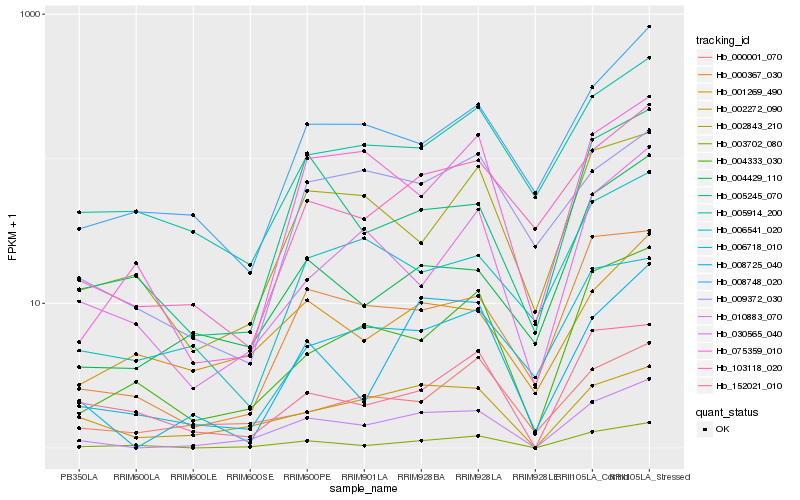
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010883_070 |
0.0 |
- |
- |
PREDICTED: biotin--protein ligase 2-like [Jatropha curcas] |
| 2 |
Hb_002272_090 |
0.1745159346 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 3 |
Hb_003702_080 |
0.1790040536 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 4 |
Hb_000367_030 |
0.1809352657 |
- |
- |
hypothetical protein POPTR_0015s06690g [Populus trichocarpa] |
| 5 |
Hb_008725_040 |
0.1822008487 |
- |
- |
PREDICTED: uncharacterized protein At4g29660 [Nelumbo nucifera] |
| 6 |
Hb_004333_030 |
0.1871614192 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 26A [Jatropha curcas] |
| 7 |
Hb_103118_020 |
0.1951224363 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 8 |
Hb_006718_010 |
0.1977314836 |
- |
- |
PREDICTED: histone H2AX-like [Elaeis guineensis] |
| 9 |
Hb_075359_010 |
0.1993955859 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 10 |
Hb_005245_070 |
0.201429926 |
- |
- |
PREDICTED: uncharacterized protein LOC105643658 [Jatropha curcas] |
| 11 |
Hb_002843_210 |
0.2045081437 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |
| 12 |
Hb_005914_200 |
0.2079654375 |
- |
- |
PREDICTED: 60S ribosomal protein L23-like isoform X1 [Tarenaya hassleriana] |
| 13 |
Hb_001269_490 |
0.2087726768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_008748_020 |
0.2088443739 |
- |
- |
hypothetical protein B456_005G210700 [Gossypium raimondii] |
| 15 |
Hb_152021_010 |
0.2112320312 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 16 |
Hb_030565_040 |
0.2114792108 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 17 |
Hb_006541_020 |
0.2119256377 |
- |
- |
unknown [Lotus japonicus] |
| 18 |
Hb_000001_070 |
0.2123511399 |
- |
- |
- |
| 19 |
Hb_004429_110 |
0.2153209416 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 20 |
Hb_009372_030 |
0.2167971785 |
- |
- |
50S ribosomal protein L6, putative [Ricinus communis] |