| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
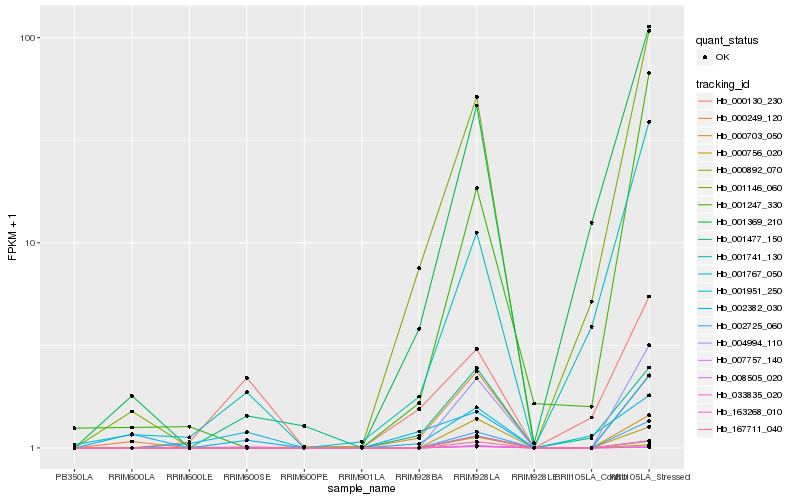
Hb_000892_070 |
0.0 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_002725_060 |
0.2310251374 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 3 |
Hb_004994_110 |
0.2313980993 |
- |
- |
- |
| 4 |
Hb_167711_040 |
0.2415061238 |
- |
- |
PREDICTED: cucumisin-like [Jatropha curcas] |
| 5 |
Hb_007757_140 |
0.2611748139 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 6 |
Hb_000130_230 |
0.2634439993 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098-like [Jatropha curcas] |
| 7 |
Hb_001951_250 |
0.2672133714 |
- |
- |
- |
| 8 |
Hb_000703_050 |
0.2720118403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000756_020 |
0.2844206259 |
- |
- |
BnaA05g19440D [Brassica napus] |
| 10 |
Hb_001477_150 |
0.2870314844 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001767_050 |
0.2959076592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001146_060 |
0.2960767589 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 13 |
Hb_001247_330 |
0.3061001032 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 14 |
Hb_163268_010 |
0.307127744 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 15 |
Hb_001741_130 |
0.3072331714 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis sativus] |
| 16 |
Hb_000249_120 |
0.3111593608 |
- |
- |
PREDICTED: uncharacterized protein LOC104613364 [Nelumbo nucifera] |
| 17 |
Hb_001369_210 |
0.3191735774 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 18 |
Hb_008505_020 |
0.3196730615 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 19 |
Hb_033835_020 |
0.3203745197 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_002382_030 |
0.3209003256 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2-like isoform X2 [Malus domestica] |