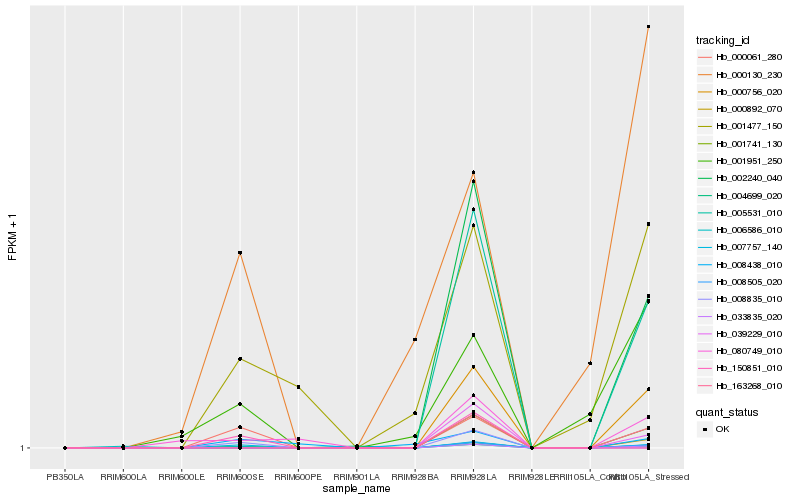
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007757_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 2 |
Hb_000892_070 |
0.2611748139 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_006586_010 |
0.2949918171 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_008438_010 |
0.3122574533 |
- |
- |
PREDICTED: uncharacterized protein LOC104245771 [Nicotiana sylvestris] |
| 5 |
Hb_001477_150 |
0.3193887481 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_080749_010 |
0.330893717 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Populus euphratica] |
| 7 |
Hb_001951_250 |
0.3423993937 |
- |
- |
- |
| 8 |
Hb_004699_020 |
0.355334672 |
- |
- |
PREDICTED: uncharacterized protein LOC103410170 [Malus domestica] |
| 9 |
Hb_008505_020 |
0.3638353123 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 10 |
Hb_033835_020 |
0.3638378452 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_005531_010 |
0.3639281654 |
- |
- |
hypothetical protein CICLE_v10033008mg [Citrus clementina] |
| 12 |
Hb_001741_130 |
0.3642441843 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis sativus] |
| 13 |
Hb_163268_010 |
0.3642514893 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 14 |
Hb_002240_040 |
0.3653529688 |
- |
- |
- |
| 15 |
Hb_000130_230 |
0.366314309 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098-like [Jatropha curcas] |
| 16 |
Hb_000756_020 |
0.3675644903 |
- |
- |
BnaA05g19440D [Brassica napus] |
| 17 |
Hb_039229_010 |
0.3807861166 |
- |
- |
- |
| 18 |
Hb_000061_280 |
0.3840114722 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g44230 [Jatropha curcas] |
| 19 |
Hb_150851_010 |
0.3867919577 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_008835_010 |
0.3891465243 |
- |
- |
PREDICTED: uncharacterized protein LOC105851721 [Cicer arietinum] |