| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
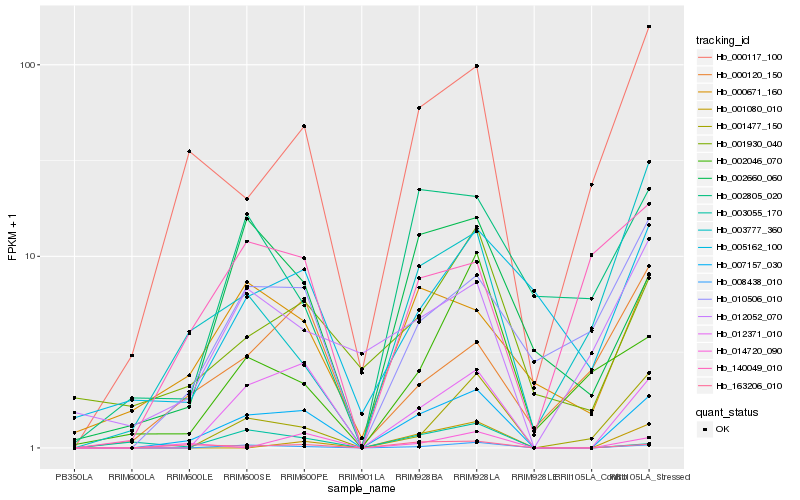
Hb_007157_030 |
0.0 |
- |
- |
PREDICTED: protein LURP-one-related 6-like [Jatropha curcas] |
| 2 |
Hb_012371_010 |
0.1795393121 |
- |
- |
PREDICTED: uncharacterized protein LOC105641615 [Jatropha curcas] |
| 3 |
Hb_008438_010 |
0.1889410923 |
- |
- |
PREDICTED: uncharacterized protein LOC104245771 [Nicotiana sylvestris] |
| 4 |
Hb_163206_010 |
0.2288241231 |
- |
- |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 5 |
Hb_002660_060 |
0.2632152471 |
- |
- |
PREDICTED: uncharacterized protein LOC105642215 [Jatropha curcas] |
| 6 |
Hb_000671_160 |
0.2714694055 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000117_100 |
0.2723623609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001930_040 |
0.2780072645 |
- |
- |
PREDICTED: serine acetyltransferase 1, chloroplastic-like [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_001477_150 |
0.2805287588 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_010506_010 |
0.2870034794 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 11 |
Hb_012052_070 |
0.3070095171 |
- |
- |
PREDICTED: uncharacterized protein LOC105635597 [Jatropha curcas] |
| 12 |
Hb_005162_100 |
0.3094870525 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_002805_020 |
0.3136775139 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_003055_170 |
0.318592394 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 25 isoform X1 [Jatropha curcas] |
| 15 |
Hb_140049_010 |
0.3193563281 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 16 |
Hb_014720_090 |
0.3204149123 |
- |
- |
- |
| 17 |
Hb_001080_010 |
0.3219662129 |
- |
- |
- |
| 18 |
Hb_000120_150 |
0.3228796243 |
- |
- |
hypothetical protein JCGZ_08453 [Jatropha curcas] |
| 19 |
Hb_003777_360 |
0.327481974 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 20 |
Hb_002046_070 |
0.3276839947 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like isoform X1 [Populus euphratica] |