| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
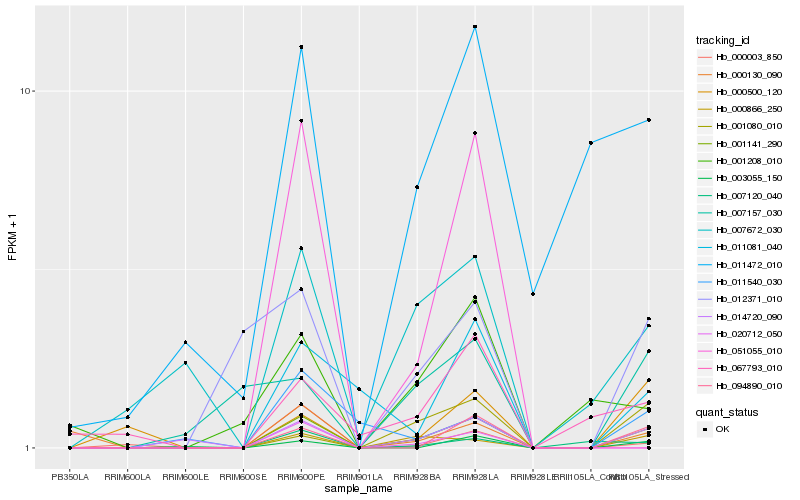
Hb_001141_290 |
0.0 |
- |
- |
- |
| 2 |
Hb_014720_090 |
0.2115978845 |
- |
- |
- |
| 3 |
Hb_000866_250 |
0.2343811091 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 4 |
Hb_094890_010 |
0.2424508732 |
- |
- |
- |
| 5 |
Hb_003055_150 |
0.2846692088 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 6 |
Hb_011081_040 |
0.3029565749 |
- |
- |
hypothetical protein JCGZ_20956 [Jatropha curcas] |
| 7 |
Hb_001080_010 |
0.3037573052 |
- |
- |
- |
| 8 |
Hb_000130_090 |
0.3046852924 |
- |
- |
Anther-specific protein LAT52 precursor, putative [Ricinus communis] |
| 9 |
Hb_012371_010 |
0.3119076317 |
- |
- |
PREDICTED: uncharacterized protein LOC105641615 [Jatropha curcas] |
| 10 |
Hb_007672_030 |
0.3132235819 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 11 |
Hb_000003_850 |
0.3190074315 |
- |
- |
PREDICTED: uncharacterized protein LOC105633597 [Jatropha curcas] |
| 12 |
Hb_007120_040 |
0.3195936647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000500_120 |
0.3202782169 |
- |
- |
PREDICTED: uncharacterized protein LOC105648659 [Jatropha curcas] |
| 14 |
Hb_011472_010 |
0.3328598215 |
- |
- |
PREDICTED: protein yippee-like [Jatropha curcas] |
| 15 |
Hb_067793_010 |
0.3371477271 |
- |
- |
PREDICTED: uncharacterized protein LOC104112577 [Nicotiana tomentosiformis] |
| 16 |
Hb_007157_030 |
0.3414802041 |
- |
- |
PREDICTED: protein LURP-one-related 6-like [Jatropha curcas] |
| 17 |
Hb_001208_010 |
0.3450609619 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF010-like [Jatropha curcas] |
| 18 |
Hb_011540_030 |
0.3475394033 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX5-like isoform X6 [Glycine max] |
| 19 |
Hb_051055_010 |
0.3501470602 |
- |
- |
- |
| 20 |
Hb_020712_050 |
0.3504293709 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |