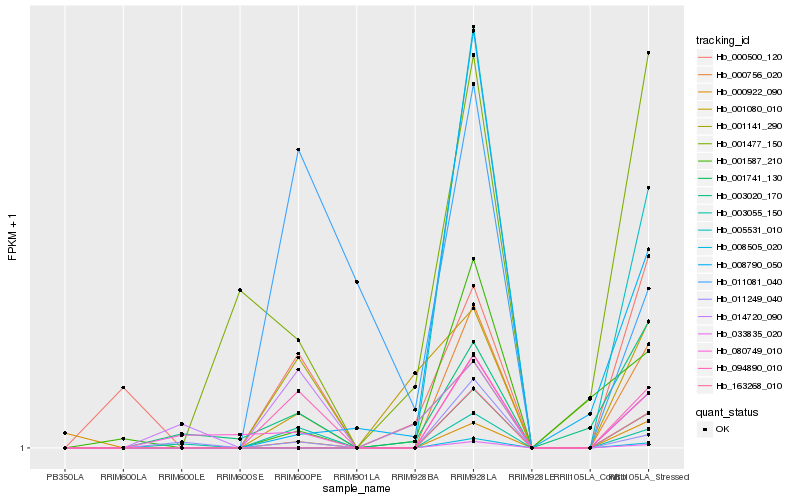
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094890_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_003055_150 |
0.1615485002 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_001141_290 |
0.2424508732 |
- |
- |
- |
| 4 |
Hb_000500_120 |
0.3076122127 |
- |
- |
PREDICTED: uncharacterized protein LOC105648659 [Jatropha curcas] |
| 5 |
Hb_014720_090 |
0.3121930827 |
- |
- |
- |
| 6 |
Hb_080749_010 |
0.3222246463 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Populus euphratica] |
| 7 |
Hb_011081_040 |
0.3254545935 |
- |
- |
hypothetical protein JCGZ_20956 [Jatropha curcas] |
| 8 |
Hb_003020_170 |
0.3362365554 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 9 |
Hb_011249_040 |
0.3522216785 |
- |
- |
PREDICTED: acetylajmalan esterase-like [Populus euphratica] |
| 10 |
Hb_001080_010 |
0.367145743 |
- |
- |
- |
| 11 |
Hb_001477_150 |
0.3746671039 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000922_090 |
0.3803184183 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g11290-like [Populus euphratica] |
| 13 |
Hb_163268_010 |
0.3834110106 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 14 |
Hb_001741_130 |
0.3834166255 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis sativus] |
| 15 |
Hb_000756_020 |
0.3837006252 |
- |
- |
BnaA05g19440D [Brassica napus] |
| 16 |
Hb_008505_020 |
0.384469208 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 17 |
Hb_033835_020 |
0.3845505763 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_005531_010 |
0.3851837207 |
- |
- |
hypothetical protein CICLE_v10033008mg [Citrus clementina] |
| 19 |
Hb_008790_050 |
0.3870456543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001587_210 |
0.387248991 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |