| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
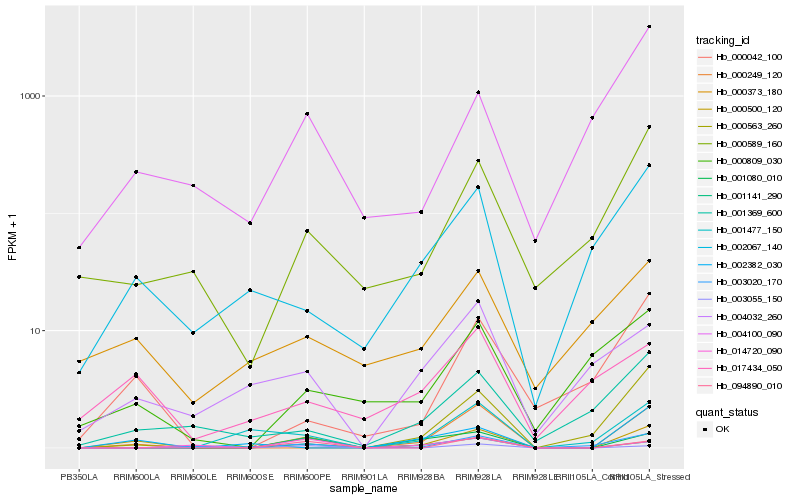
Hb_000500_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648659 [Jatropha curcas] |
| 2 |
Hb_094890_010 |
0.3076122127 |
- |
- |
- |
| 3 |
Hb_001141_290 |
0.3202782169 |
- |
- |
- |
| 4 |
Hb_001080_010 |
0.3209093921 |
- |
- |
- |
| 5 |
Hb_000563_260 |
0.3244681557 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 6 |
Hb_000042_100 |
0.3390099448 |
- |
- |
PREDICTED: uncharacterized protein LOC104448410 [Eucalyptus grandis] |
| 7 |
Hb_000249_120 |
0.3408702835 |
- |
- |
PREDICTED: uncharacterized protein LOC104613364 [Nelumbo nucifera] |
| 8 |
Hb_000589_160 |
0.3485630111 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 9 |
Hb_003020_170 |
0.3505795193 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 10 |
Hb_003055_150 |
0.3530156167 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 11 |
Hb_002382_030 |
0.3608653857 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2-like isoform X2 [Malus domestica] |
| 12 |
Hb_002067_140 |
0.3626829572 |
- |
- |
- |
| 13 |
Hb_017434_050 |
0.3644524642 |
- |
- |
PREDICTED: importin subunit beta-3-like [Jatropha curcas] |
| 14 |
Hb_014720_090 |
0.3649328534 |
- |
- |
- |
| 15 |
Hb_004100_090 |
0.366357994 |
- |
- |
RecName: Full=Profilin-6; AltName: Full=Pollen allergen Hev b 8.0204; AltName: Allergen=Hev b 8.0204 [Hevea brasiliensis] |
| 16 |
Hb_001369_600 |
0.3671969477 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_000809_030 |
0.3685003944 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At5g66900 [Prunus mume] |
| 18 |
Hb_004032_260 |
0.369482104 |
- |
- |
PREDICTED: uncharacterized protein LOC105635147 [Jatropha curcas] |
| 19 |
Hb_001477_150 |
0.3716915258 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000373_180 |
0.3717956467 |
transcription factor |
TF Family: Trihelix |
hypothetical protein JCGZ_25182 [Jatropha curcas] |