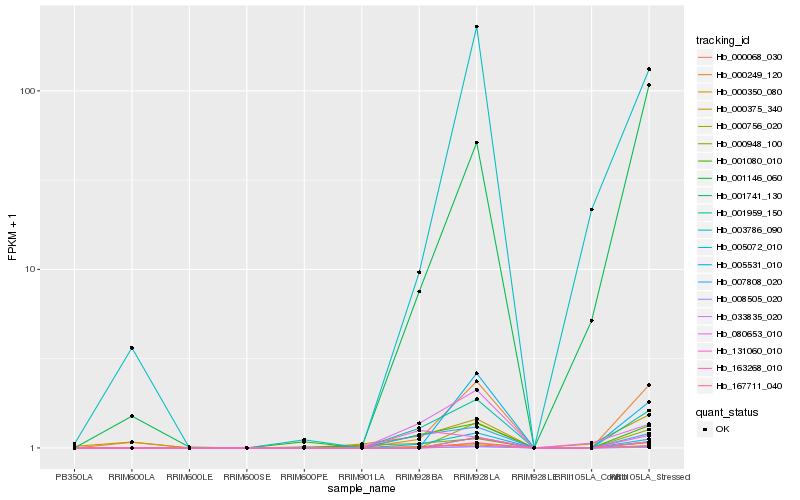
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_005072_010 |
0.0626881277 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 3 |
Hb_000068_030 |
0.0713188794 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 4 |
Hb_000350_080 |
0.0803179323 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 5 |
Hb_007808_020 |
0.1031058687 |
- |
- |
- |
| 6 |
Hb_080653_010 |
0.1967661764 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_000249_120 |
0.2118757666 |
- |
- |
PREDICTED: uncharacterized protein LOC104613364 [Nelumbo nucifera] |
| 8 |
Hb_001080_010 |
0.218060616 |
- |
- |
- |
| 9 |
Hb_000948_100 |
0.2630212315 |
- |
- |
- |
| 10 |
Hb_131060_010 |
0.2712300207 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 11 |
Hb_003786_090 |
0.2762885842 |
- |
- |
hypothetical protein JCGZ_19199 [Jatropha curcas] |
| 12 |
Hb_000756_020 |
0.2935439148 |
- |
- |
BnaA05g19440D [Brassica napus] |
| 13 |
Hb_163268_010 |
0.2964671299 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 14 |
Hb_001741_130 |
0.2964895274 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis sativus] |
| 15 |
Hb_000375_340 |
0.2983660508 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Populus euphratica] |
| 16 |
Hb_008505_020 |
0.2996206181 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 17 |
Hb_033835_020 |
0.299824554 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_001146_060 |
0.301121214 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 19 |
Hb_005531_010 |
0.3013319035 |
- |
- |
hypothetical protein CICLE_v10033008mg [Citrus clementina] |
| 20 |
Hb_167711_040 |
0.3056146677 |
- |
- |
PREDICTED: cucumisin-like [Jatropha curcas] |