| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
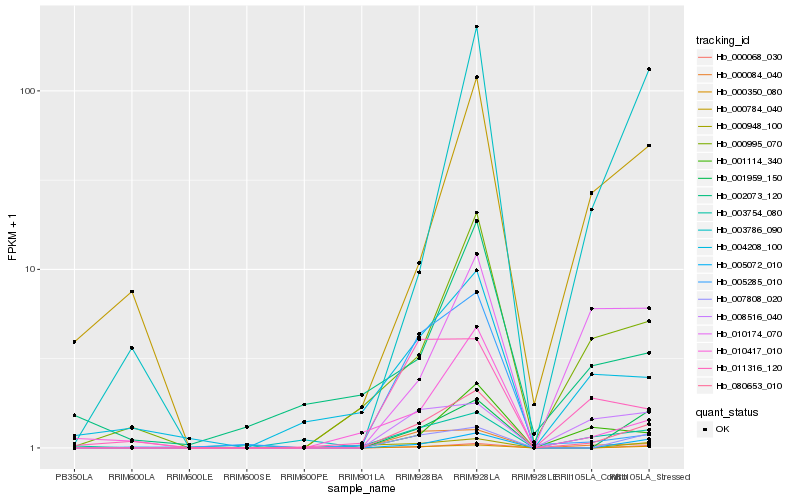
Hb_080653_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000350_080 |
0.1570234924 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 3 |
Hb_000068_030 |
0.1830720959 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 4 |
Hb_005072_010 |
0.1839330574 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 5 |
Hb_007808_020 |
0.187571644 |
- |
- |
- |
| 6 |
Hb_001959_150 |
0.1967661764 |
- |
- |
- |
| 7 |
Hb_000084_040 |
0.2155555575 |
- |
- |
hypothetical protein RCOM_1007940 [Ricinus communis] |
| 8 |
Hb_001114_340 |
0.2299420612 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 9 |
Hb_000995_070 |
0.2336438997 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 10 |
Hb_003754_080 |
0.2463959003 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 11 |
Hb_010417_010 |
0.2627620208 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_004208_100 |
0.2678402088 |
- |
- |
Ribosomal protein L3 plastid isoform 1 [Theobroma cacao] |
| 13 |
Hb_008516_040 |
0.2722601381 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003786_090 |
0.2780766014 |
- |
- |
hypothetical protein JCGZ_19199 [Jatropha curcas] |
| 15 |
Hb_011316_120 |
0.2848987096 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH118-like [Jatropha curcas] |
| 16 |
Hb_000784_040 |
0.2849648499 |
- |
- |
hypothetical protein POPTR_0013s11550g [Populus trichocarpa] |
| 17 |
Hb_005285_010 |
0.2912587473 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_010174_070 |
0.291443572 |
- |
- |
- |
| 19 |
Hb_000948_100 |
0.2964911207 |
- |
- |
- |
| 20 |
Hb_002073_120 |
0.3007467563 |
- |
- |
Adenine nucleotide alpha hydrolases-like superfamily protein [Theobroma cacao] |