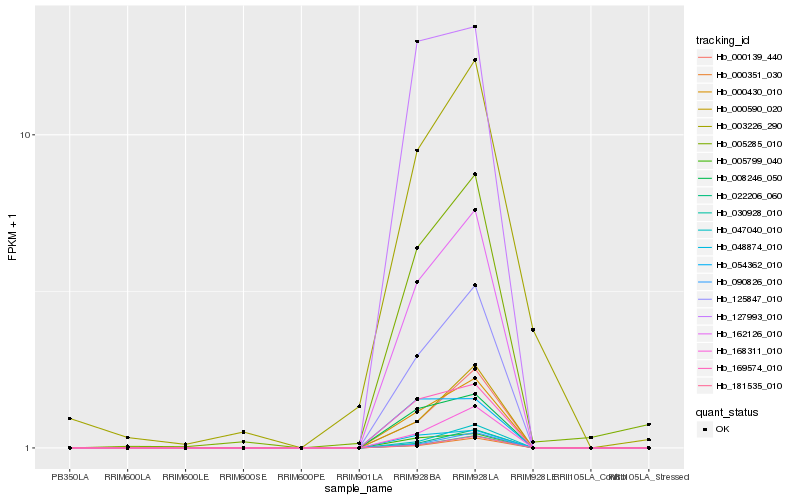
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005285_010 |
0.0 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_162126_010 |
0.1432511978 |
- |
- |
- |
| 3 |
Hb_000430_010 |
0.1453859571 |
- |
- |
- |
| 4 |
Hb_125847_010 |
0.1490422458 |
- |
- |
- |
| 5 |
Hb_008246_050 |
0.1536550596 |
- |
- |
- |
| 6 |
Hb_005799_040 |
0.1536716922 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4-like isoform X1 [Gossypium raimondii] |
| 7 |
Hb_048874_010 |
0.1560401969 |
- |
- |
hypothetical protein POPTR_0016s10870g [Populus trichocarpa] |
| 8 |
Hb_169574_010 |
0.1570601313 |
- |
- |
PREDICTED: uncharacterized protein LOC104120373 [Nicotiana tomentosiformis] |
| 9 |
Hb_168311_010 |
0.1735752959 |
- |
- |
- |
| 10 |
Hb_127993_010 |
0.1812741874 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X4 [Jatropha curcas] |
| 11 |
Hb_003226_290 |
0.1844040321 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_000139_440 |
0.1871551731 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_000590_020 |
0.1941756609 |
- |
- |
hypothetical protein JCGZ_18748 [Jatropha curcas] |
| 14 |
Hb_054362_010 |
0.1957343108 |
- |
- |
PREDICTED: uncharacterized protein LOC105650428 [Jatropha curcas] |
| 15 |
Hb_047040_010 |
0.1972809278 |
- |
- |
integrase [Gossypium herbaceum] |
| 16 |
Hb_022206_060 |
0.2029600701 |
- |
- |
- |
| 17 |
Hb_090826_010 |
0.2060189673 |
- |
- |
PREDICTED: uncharacterized protein LOC105786910 [Gossypium raimondii] |
| 18 |
Hb_030928_010 |
0.2093714121 |
- |
- |
- |
| 19 |
Hb_181535_010 |
0.2098691602 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_000351_030 |
0.211570724 |
- |
- |
putative NBS-LRR type disease resistance protein [Prunus persica] |