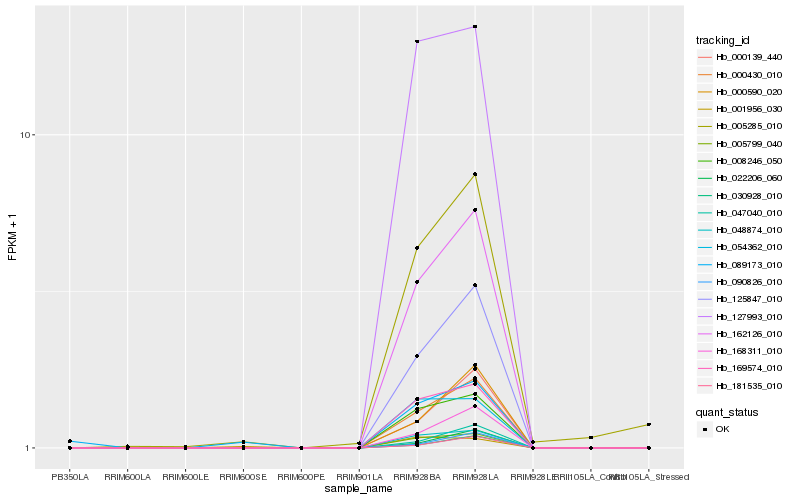
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005799_040 |
0.0 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4-like isoform X1 [Gossypium raimondii] |
| 2 |
Hb_008246_050 |
4.61958e-05 |
- |
- |
- |
| 3 |
Hb_048874_010 |
0.0062797337 |
- |
- |
hypothetical protein POPTR_0016s10870g [Populus trichocarpa] |
| 4 |
Hb_169574_010 |
0.008826202 |
- |
- |
PREDICTED: uncharacterized protein LOC104120373 [Nicotiana tomentosiformis] |
| 5 |
Hb_127993_010 |
0.0559908633 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X4 [Jatropha curcas] |
| 6 |
Hb_162126_010 |
0.0632038542 |
- |
- |
- |
| 7 |
Hb_054362_010 |
0.0785841086 |
- |
- |
PREDICTED: uncharacterized protein LOC105650428 [Jatropha curcas] |
| 8 |
Hb_000430_010 |
0.0824882532 |
- |
- |
- |
| 9 |
Hb_125847_010 |
0.0985920704 |
- |
- |
- |
| 10 |
Hb_005285_010 |
0.1536716922 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_168311_010 |
0.1554405539 |
- |
- |
- |
| 12 |
Hb_000139_440 |
0.1778655625 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_000590_020 |
0.188499969 |
- |
- |
hypothetical protein JCGZ_18748 [Jatropha curcas] |
| 14 |
Hb_047040_010 |
0.193051126 |
- |
- |
integrase [Gossypium herbaceum] |
| 15 |
Hb_022206_060 |
0.2011710846 |
- |
- |
- |
| 16 |
Hb_090826_010 |
0.2054478862 |
- |
- |
PREDICTED: uncharacterized protein LOC105786910 [Gossypium raimondii] |
| 17 |
Hb_030928_010 |
0.2100657914 |
- |
- |
- |
| 18 |
Hb_089173_010 |
0.2103623738 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 19 |
Hb_181535_010 |
0.210745623 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_001956_030 |
0.2109872105 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Populus euphratica] |