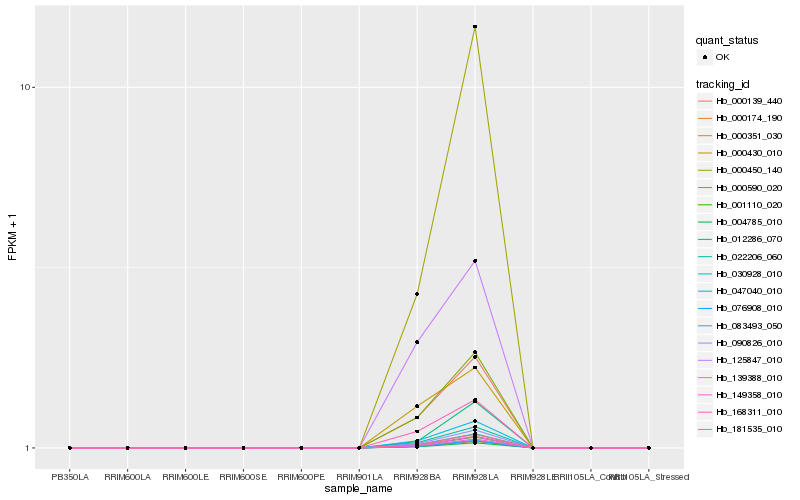
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000590_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_18748 [Jatropha curcas] |
| 2 |
Hb_047040_010 |
0.0046430457 |
- |
- |
integrase [Gossypium herbaceum] |
| 3 |
Hb_000139_440 |
0.0108277543 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_022206_060 |
0.01294105 |
- |
- |
- |
| 5 |
Hb_090826_010 |
0.0173190872 |
- |
- |
PREDICTED: uncharacterized protein LOC105786910 [Gossypium raimondii] |
| 6 |
Hb_030928_010 |
0.0220522343 |
- |
- |
- |
| 7 |
Hb_181535_010 |
0.0227495594 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 8 |
Hb_000351_030 |
0.0251233402 |
- |
- |
putative NBS-LRR type disease resistance protein [Prunus persica] |
| 9 |
Hb_139388_010 |
0.0254396818 |
- |
- |
PREDICTED: uncharacterized protein LOC104222982, partial [Nicotiana sylvestris] |
| 10 |
Hb_001110_020 |
0.0259692433 |
- |
- |
hypothetical protein JCGZ_03693 [Jatropha curcas] |
| 11 |
Hb_149358_010 |
0.0282179669 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 12 |
Hb_083493_050 |
0.0295996466 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 13 |
Hb_076908_010 |
0.0301220521 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 14 |
Hb_000174_190 |
0.0308462896 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_168311_010 |
0.0335677575 |
- |
- |
- |
| 16 |
Hb_004785_010 |
0.0349654169 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 17 |
Hb_125847_010 |
0.0907190885 |
- |
- |
- |
| 18 |
Hb_000430_010 |
0.1067955777 |
- |
- |
- |
| 19 |
Hb_012286_070 |
0.1114618763 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 20 |
Hb_000450_140 |
0.1115598803 |
- |
- |
hypothetical protein POPTR_0009s06810g [Populus trichocarpa] |