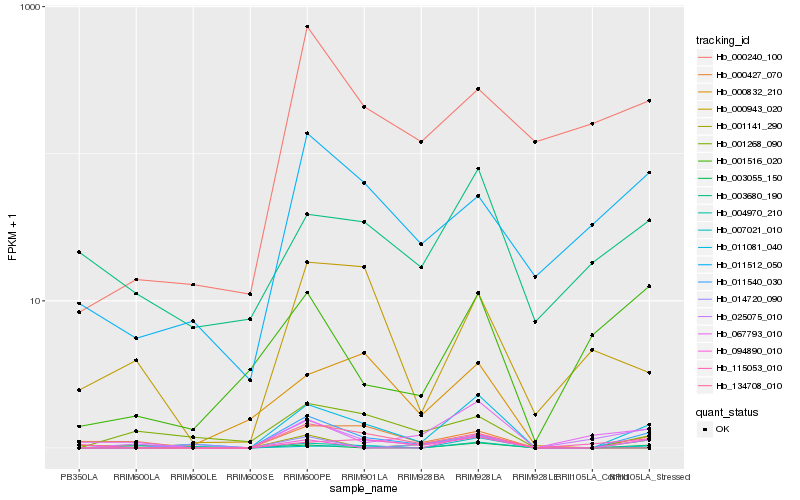
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011081_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_20956 [Jatropha curcas] |
| 2 |
Hb_011540_030 |
0.2774983636 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX5-like isoform X6 [Glycine max] |
| 3 |
Hb_001141_290 |
0.3029565749 |
- |
- |
- |
| 4 |
Hb_094890_010 |
0.3254545935 |
- |
- |
- |
| 5 |
Hb_134708_010 |
0.3266962943 |
- |
- |
PREDICTED: uncharacterized protein LOC102604542 [Solanum tuberosum] |
| 6 |
Hb_000427_070 |
0.331058202 |
- |
- |
- |
| 7 |
Hb_067793_010 |
0.343794836 |
- |
- |
PREDICTED: uncharacterized protein LOC104112577 [Nicotiana tomentosiformis] |
| 8 |
Hb_003055_150 |
0.3502337358 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 9 |
Hb_115053_010 |
0.3614806167 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 10 |
Hb_014720_090 |
0.3621226774 |
- |
- |
- |
| 11 |
Hb_004970_210 |
0.3638225486 |
- |
- |
- |
| 12 |
Hb_001268_090 |
0.3702776532 |
- |
- |
Uncharacterized protein TCM_020504 [Theobroma cacao] |
| 13 |
Hb_025075_010 |
0.3728754214 |
- |
- |
50S ribosomal protein L24 [Glycine soja] |
| 14 |
Hb_000240_100 |
0.3735537457 |
- |
- |
PREDICTED: ras-related protein Rab5-like [Eucalyptus grandis] |
| 15 |
Hb_011512_050 |
0.3768724115 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 13-A isoform 2 [Theobroma cacao] |
| 16 |
Hb_007021_010 |
0.3849920209 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0939100 [Ricinus communis] |
| 17 |
Hb_000943_020 |
0.3854431631 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 18 |
Hb_000832_210 |
0.3935629498 |
- |
- |
PREDICTED: WAT1-related protein At2g39510 [Jatropha curcas] |
| 19 |
Hb_003680_190 |
0.3936853046 |
- |
- |
hypothetical protein POPTR_0006s11880g [Populus trichocarpa] |
| 20 |
Hb_001516_020 |
0.394000886 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-9-like isoform X2 [Jatropha curcas] |