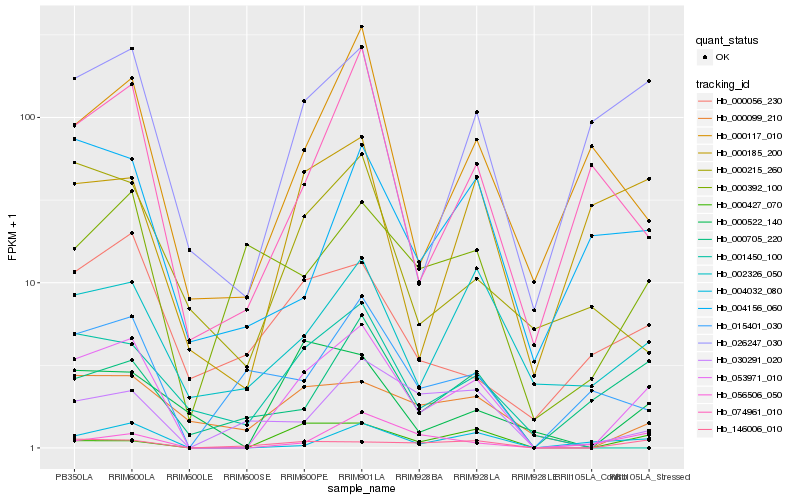
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_053971_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_004032_080 |
0.2464950118 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL11 [Jatropha curcas] |
| 3 |
Hb_026247_030 |
0.2577106362 |
- |
- |
PREDICTED: protein trichome birefringence-like 42 [Jatropha curcas] |
| 4 |
Hb_146006_010 |
0.2650009236 |
- |
- |
PREDICTED: uncharacterized protein LOC104091534 [Nicotiana tomentosiformis] |
| 5 |
Hb_002326_050 |
0.2681751355 |
- |
- |
PREDICTED: uncharacterized protein LOC105633701 [Jatropha curcas] |
| 6 |
Hb_000522_140 |
0.2753241328 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 7 |
Hb_000099_210 |
0.2757594212 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |
| 8 |
Hb_074961_010 |
0.2785020029 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 9 |
Hb_000427_070 |
0.280592131 |
- |
- |
- |
| 10 |
Hb_000185_200 |
0.2823679732 |
- |
- |
PREDICTED: protein trichome birefringence-like 42 [Jatropha curcas] |
| 11 |
Hb_000705_220 |
0.2823692034 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_030291_020 |
0.283591019 |
- |
- |
PREDICTED: protein argonaute 5 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001450_100 |
0.2850511878 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 14 |
Hb_000392_100 |
0.2872241215 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_015401_030 |
0.2876548741 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 16 |
Hb_000117_010 |
0.2893003544 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like [Malus domestica] |
| 17 |
Hb_000215_260 |
0.2894136524 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: leucine-rich repeat extensin-like protein 4 [Jatropha curcas] |
| 18 |
Hb_000056_230 |
0.2918264561 |
transcription factor |
TF Family: TCP |
hypothetical protein JCGZ_15588 [Jatropha curcas] |
| 19 |
Hb_004156_060 |
0.2928324243 |
- |
- |
neutral/alkaline invertase 1 [Hevea brasiliensis] |
| 20 |
Hb_056506_050 |
0.293405495 |
- |
- |
hypothetical protein B456_007G294000 [Gossypium raimondii] |