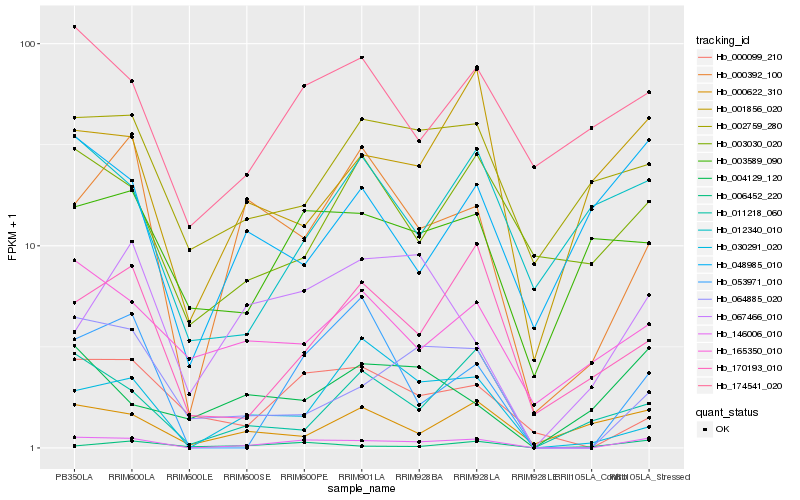
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146006_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104091534 [Nicotiana tomentosiformis] |
| 2 |
Hb_000099_210 |
0.2542184689 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |
| 3 |
Hb_064885_020 |
0.2586996406 |
- |
- |
- |
| 4 |
Hb_000392_100 |
0.2641439737 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_053971_010 |
0.2650009236 |
- |
- |
- |
| 6 |
Hb_174541_020 |
0.2774310549 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 6-like [Populus euphratica] |
| 7 |
Hb_003589_090 |
0.2804633754 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |
| 8 |
Hb_001856_020 |
0.2806836189 |
transcription factor |
TF Family: Orphans |
ethylene receptor, putative [Ricinus communis] |
| 9 |
Hb_170193_010 |
0.2831975381 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 10 |
Hb_011218_060 |
0.2843220426 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 11 |
Hb_165350_010 |
0.2891148185 |
- |
- |
unnamed protein product [Triticum aestivum] |
| 12 |
Hb_000622_310 |
0.2900016477 |
- |
- |
PREDICTED: BON1-associated protein 2 [Jatropha curcas] |
| 13 |
Hb_006452_220 |
0.2902366047 |
- |
- |
PREDICTED: uncharacterized protein LOC105635043 [Jatropha curcas] |
| 14 |
Hb_002759_280 |
0.2908254692 |
- |
- |
PREDICTED: uncharacterized protein LOC105649824 [Jatropha curcas] |
| 15 |
Hb_067466_010 |
0.2909807833 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 16 |
Hb_003030_020 |
0.2915694985 |
- |
- |
PREDICTED: uncharacterized protein LOC105632544 [Jatropha curcas] |
| 17 |
Hb_030291_020 |
0.2938220709 |
- |
- |
PREDICTED: protein argonaute 5 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004129_120 |
0.2971255707 |
- |
- |
- |
| 19 |
Hb_048985_010 |
0.2977964802 |
- |
- |
- |
| 20 |
Hb_012340_010 |
0.2987329162 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Jatropha curcas] |