| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
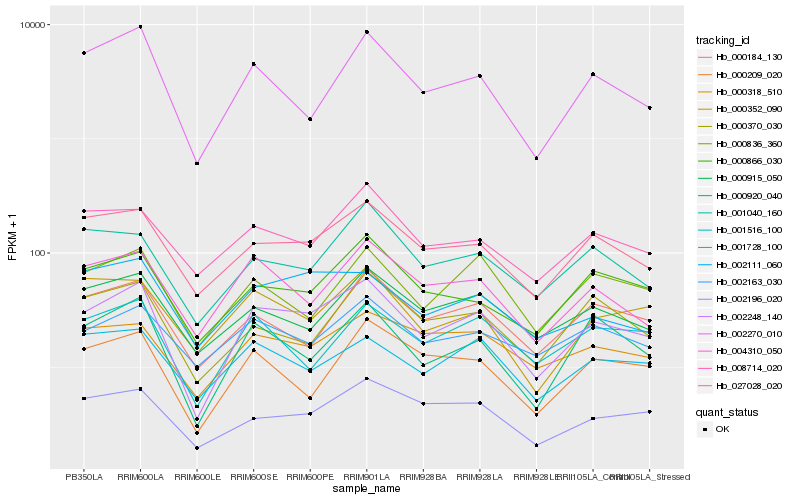
Hb_002248_140 |
0.0 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 2 |
Hb_027028_020 |
0.1041442963 |
- |
- |
PREDICTED: uncharacterized protein LOC105633192 [Jatropha curcas] |
| 3 |
Hb_000920_040 |
0.1117343225 |
- |
- |
PREDICTED: F-box protein At1g30790-like [Jatropha curcas] |
| 4 |
Hb_000866_030 |
0.1213301421 |
- |
- |
hypothetical protein RCOM_1615820 [Ricinus communis] |
| 5 |
Hb_004310_050 |
0.1231391267 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 6 |
Hb_000915_050 |
0.1242812635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002196_020 |
0.1263591049 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_008714_020 |
0.1269737591 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 9 |
Hb_000184_130 |
0.1278172072 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002270_010 |
0.1280632284 |
- |
- |
Chaperone protein dnaJ, putative [Ricinus communis] |
| 11 |
Hb_000209_020 |
0.1283959888 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |
| 12 |
Hb_000318_510 |
0.1299165749 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 13 |
Hb_002163_030 |
0.1300628309 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 59 kDa regulatory subunit B' gamma isoform [Jatropha curcas] |
| 14 |
Hb_000836_360 |
0.1328820681 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001516_100 |
0.1335267792 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_002111_060 |
0.1341529306 |
- |
- |
PREDICTED: toll-like receptor 13 [Jatropha curcas] |
| 17 |
Hb_001040_160 |
0.1343175722 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 18 |
Hb_000370_030 |
0.1344340115 |
- |
- |
PREDICTED: uncharacterized protein LOC105639493 [Jatropha curcas] |
| 19 |
Hb_000352_090 |
0.1351916778 |
- |
- |
PREDICTED: syntaxin-22-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001728_100 |
0.1358188376 |
- |
- |
hypothetical protein JCGZ_03429 [Jatropha curcas] |