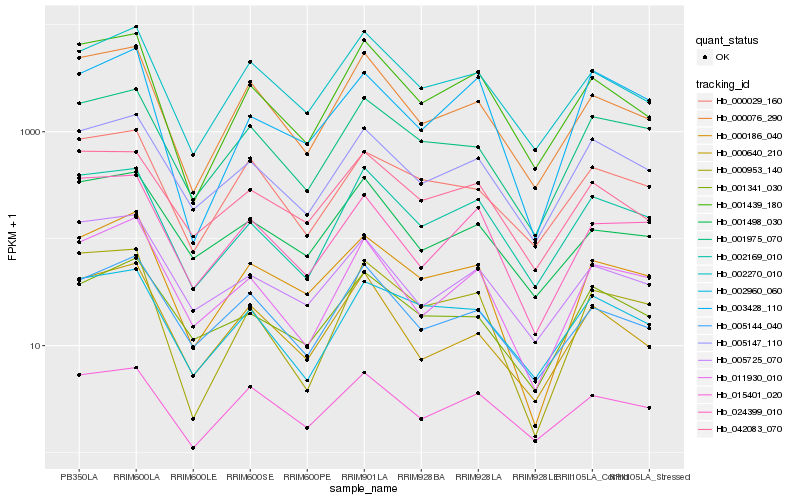
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002270_010 |
0.1074452137 |
- |
- |
Chaperone protein dnaJ, putative [Ricinus communis] |
| 3 |
Hb_001975_070 |
0.1079073308 |
- |
- |
eukaryotic translation elongation factor 1B alpha-subunit [Hevea brasiliensis] |
| 4 |
Hb_011930_010 |
0.1095128954 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_005147_110 |
0.1128592441 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 6 |
Hb_000076_290 |
0.1132535211 |
- |
- |
translation factor sui1, putative [Ricinus communis] |
| 7 |
Hb_001341_030 |
0.1133577505 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 8 |
Hb_005144_040 |
0.1181149687 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 9 |
Hb_015401_020 |
0.1190920275 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 10 |
Hb_001439_180 |
0.1217535623 |
- |
- |
hypothetical protein POPTR_0004s13090g [Populus trichocarpa] |
| 11 |
Hb_024399_010 |
0.1222278202 |
- |
- |
PREDICTED: uncharacterized protein LOC105649031 [Jatropha curcas] |
| 12 |
Hb_000029_160 |
0.1244289842 |
- |
- |
Nitrilase, putative [Ricinus communis] |
| 13 |
Hb_005725_070 |
0.1274936842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001498_030 |
0.1280680755 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit E [Populus euphratica] |
| 15 |
Hb_042083_070 |
0.1290605082 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 16 |
Hb_003428_110 |
0.1293329998 |
- |
- |
ARF2-B: ADP-ribosylation factor 2-B [Gossypium arboreum] |
| 17 |
Hb_002960_060 |
0.1302305895 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 28 [Jatropha curcas] |
| 18 |
Hb_000640_210 |
0.1313743166 |
- |
- |
hypothetical protein JCGZ_14045 [Jatropha curcas] |
| 19 |
Hb_002169_010 |
0.1317008703 |
- |
- |
PREDICTED: la-related protein 6B [Jatropha curcas] |
| 20 |
Hb_000953_140 |
0.1333125325 |
transcription factor |
TF Family: Trihelix |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |