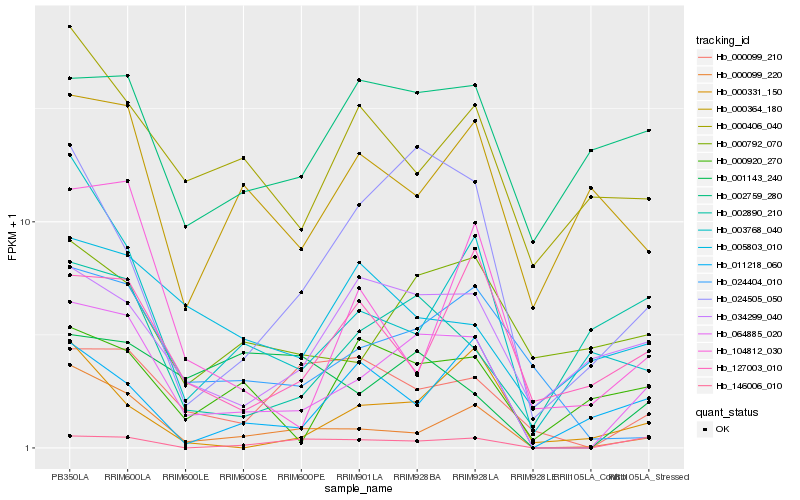
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_064885_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000099_220 |
0.2086027432 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_024505_050 |
0.2364023143 |
transcription factor |
TF Family: ERF |
PPLZ02 family protein [Populus trichocarpa] |
| 4 |
Hb_024404_010 |
0.2454441129 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Vitis vinifera] |
| 5 |
Hb_034299_040 |
0.2515993054 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 6 |
Hb_000792_070 |
0.252925563 |
- |
- |
Ataxin-3, putative [Ricinus communis] |
| 7 |
Hb_000099_210 |
0.255751679 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |
| 8 |
Hb_146006_010 |
0.2586996406 |
- |
- |
PREDICTED: uncharacterized protein LOC104091534 [Nicotiana tomentosiformis] |
| 9 |
Hb_003768_040 |
0.265240735 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_000920_270 |
0.2673116947 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 11 |
Hb_002759_280 |
0.2690379246 |
- |
- |
PREDICTED: uncharacterized protein LOC105649824 [Jatropha curcas] |
| 12 |
Hb_000406_040 |
0.2713470125 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: FRIGIDA-like protein 5 [Jatropha curcas] |
| 13 |
Hb_104812_030 |
0.2745631135 |
- |
- |
hypothetical protein POPTR_0178s00210g [Populus trichocarpa] |
| 14 |
Hb_127003_010 |
0.2746154687 |
- |
- |
PREDICTED: uncharacterized protein LOC105633613 [Jatropha curcas] |
| 15 |
Hb_000364_180 |
0.2760211019 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130 [Jatropha curcas] |
| 16 |
Hb_011218_060 |
0.2775491015 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 17 |
Hb_005803_010 |
0.2795327944 |
- |
- |
PREDICTED: probable zinc metallopeptidase EGY3, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000331_150 |
0.2797714698 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_002890_210 |
0.280021943 |
- |
- |
PREDICTED: vinorine synthase-like [Jatropha curcas] |
| 20 |
Hb_001143_240 |
0.2821068699 |
- |
- |
8-oxoguanine DNA glycosylase, putative [Ricinus communis] |