| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
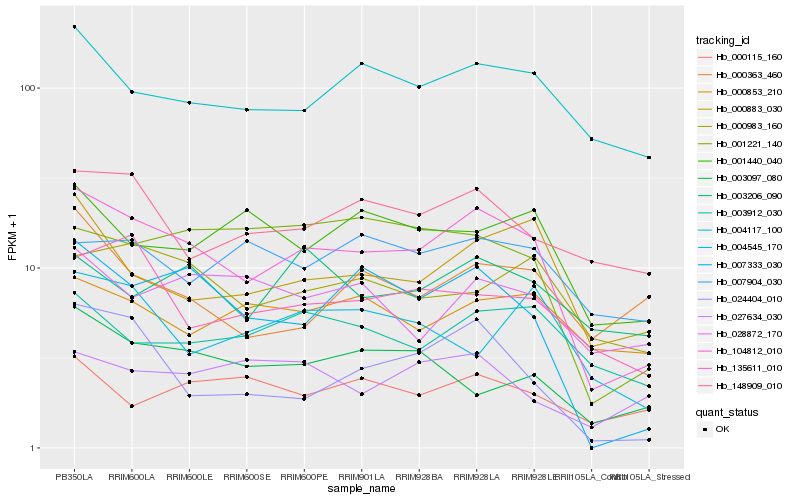
Hb_104812_010 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 9 [Populus euphratica] |
| 2 |
Hb_007333_030 |
0.1434557683 |
- |
- |
- |
| 3 |
Hb_004117_100 |
0.1552501078 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000883_030 |
0.1574166715 |
- |
- |
PREDICTED: uncharacterized protein LOC105636683 [Jatropha curcas] |
| 5 |
Hb_000983_160 |
0.1656213872 |
- |
- |
hypothetical protein B456_009G199800 [Gossypium raimondii] |
| 6 |
Hb_024404_010 |
0.1676678667 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Vitis vinifera] |
| 7 |
Hb_135611_010 |
0.167846709 |
- |
- |
PREDICTED: uncharacterized protein LOC105142085 [Populus euphratica] |
| 8 |
Hb_148909_010 |
0.1699279403 |
- |
- |
PREDICTED: uncharacterized protein LOC105638189 isoform X3 [Jatropha curcas] |
| 9 |
Hb_003912_030 |
0.1738274914 |
- |
- |
PREDICTED: uncharacterized protein LOC105638204 [Jatropha curcas] |
| 10 |
Hb_001440_040 |
0.175358913 |
- |
- |
hypothetical protein POPTR_0008s03520g [Populus trichocarpa] |
| 11 |
Hb_001221_140 |
0.1760582668 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_003206_090 |
0.1817716555 |
- |
- |
PREDICTED: protein trichome birefringence-like 5 [Jatropha curcas] |
| 13 |
Hb_028872_170 |
0.1819687595 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003097_080 |
0.1844066914 |
- |
- |
hypothetical protein JCGZ_24481 [Jatropha curcas] |
| 15 |
Hb_000853_210 |
0.1859558314 |
- |
- |
PREDICTED: uncharacterized protein LOC105642575 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007904_030 |
0.1886773282 |
- |
- |
PREDICTED: nuclear mitotic apparatus protein 1 [Jatropha curcas] |
| 17 |
Hb_000363_460 |
0.1909477916 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_004545_170 |
0.1923801171 |
- |
- |
PREDICTED: autophagy-related protein 18h [Jatropha curcas] |
| 19 |
Hb_000115_160 |
0.1926971589 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 20 |
Hb_027634_030 |
0.1928607217 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |