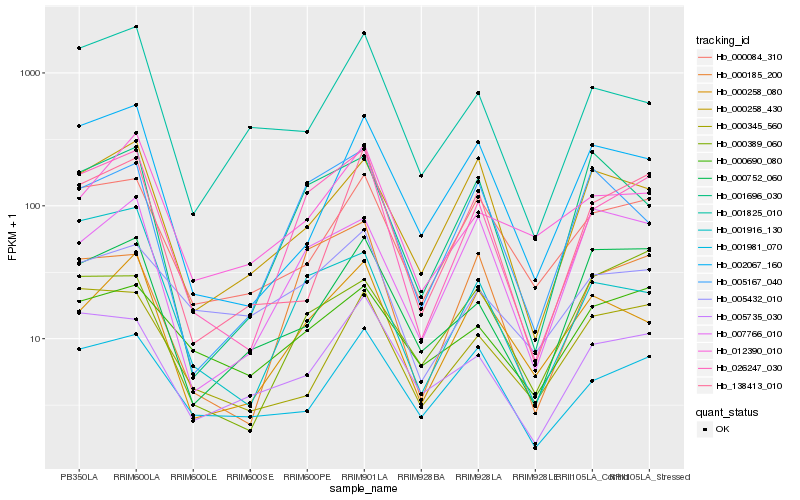
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026247_030 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence-like 42 [Jatropha curcas] |
| 2 |
Hb_000185_200 |
0.1198063618 |
- |
- |
PREDICTED: protein trichome birefringence-like 42 [Jatropha curcas] |
| 3 |
Hb_005735_030 |
0.1647729199 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000258_080 |
0.1669585079 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 5 |
Hb_000084_310 |
0.1698445779 |
- |
- |
PREDICTED: rho GTPase-activating protein 4-like [Jatropha curcas] |
| 6 |
Hb_001916_130 |
0.1741178585 |
- |
- |
Uncharacterized protein TCM_002409 [Theobroma cacao] |
| 7 |
Hb_005167_040 |
0.1745221207 |
- |
- |
PREDICTED: phospholipase A1-IIdelta [Jatropha curcas] |
| 8 |
Hb_138413_010 |
0.1753106515 |
- |
- |
hypothetical protein JCGZ_09524 [Jatropha curcas] |
| 9 |
Hb_001696_030 |
0.1762821019 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_007766_010 |
0.1776266435 |
- |
- |
hypothetical protein JCGZ_06718 [Jatropha curcas] |
| 11 |
Hb_005432_010 |
0.1805332561 |
- |
- |
- |
| 12 |
Hb_000258_430 |
0.1814492896 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 13 |
Hb_000752_060 |
0.1818332058 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 14 |
Hb_001981_070 |
0.1842420242 |
- |
- |
tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001825_010 |
0.1874001681 |
- |
- |
PREDICTED: bet1-like SNARE 1-1 [Jatropha curcas] |
| 16 |
Hb_000690_080 |
0.187816437 |
transcription factor |
TF Family: HB |
- |
| 17 |
Hb_012390_010 |
0.1888425374 |
- |
- |
pectate lyase 1 [Hevea brasiliensis] |
| 18 |
Hb_002067_160 |
0.1888667976 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 19 |
Hb_000345_560 |
0.1906664632 |
- |
- |
- |
| 20 |
Hb_000389_060 |
0.1906746856 |
- |
- |
PREDICTED: synaptotagmin-3 isoform X1 [Jatropha curcas] |