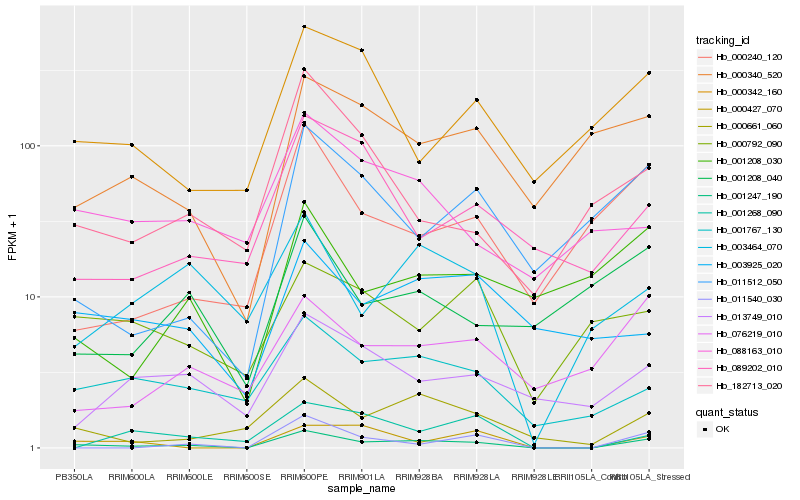
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_190 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000661_060 |
0.2479798138 |
- |
- |
hypothetical protein JCGZ_06411 [Jatropha curcas] |
| 3 |
Hb_011540_030 |
0.2717147512 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX5-like isoform X6 [Glycine max] |
| 4 |
Hb_001767_130 |
0.2741763881 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 5 |
Hb_000427_070 |
0.2830179968 |
- |
- |
- |
| 6 |
Hb_003464_070 |
0.2949521298 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 7 |
Hb_000240_120 |
0.2966476889 |
- |
- |
ras-related protein Rab-5B [Reticulomyxa filosa] |
| 8 |
Hb_076219_010 |
0.2976126428 |
- |
- |
PREDICTED: RNA pseudouridine synthase 4, mitochondrial [Jatropha curcas] |
| 9 |
Hb_011512_050 |
0.2984289148 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 13-A isoform 2 [Theobroma cacao] |
| 10 |
Hb_000340_520 |
0.3016812331 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 11 |
Hb_182713_020 |
0.3056848431 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727 [Setaria italica] |
| 12 |
Hb_013749_010 |
0.308328702 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000342_160 |
0.3086021826 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 14 |
Hb_001208_030 |
0.3088317841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_089202_010 |
0.3131706996 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Nicotiana sylvestris] |
| 16 |
Hb_088163_010 |
0.3140624379 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 17 |
Hb_003925_020 |
0.3141242587 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 18 |
Hb_001268_090 |
0.314300042 |
- |
- |
Uncharacterized protein TCM_020504 [Theobroma cacao] |
| 19 |
Hb_000792_090 |
0.3208477889 |
- |
- |
3-beta-hydroxy-delta5-steroid dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_001208_040 |
0.3210045517 |
- |
- |
hypothetical protein CISIN_1g025613mg [Citrus sinensis] |