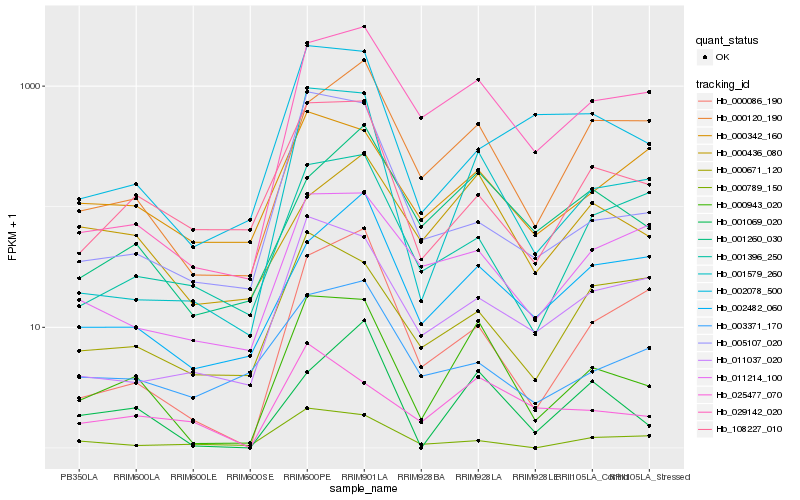
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000943_020 |
0.0 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 2 |
Hb_001579_260 |
0.1869339474 |
- |
- |
putative ADP-ribolylation factor [Arabidopsis thaliana] |
| 3 |
Hb_029142_020 |
0.2260720593 |
- |
- |
hypothetical protein POPTR_0004s23900g [Populus trichocarpa] |
| 4 |
Hb_108227_010 |
0.2296948782 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 5 |
Hb_001069_020 |
0.2319063655 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_025477_070 |
0.2353173063 |
- |
- |
hypothetical protein CICLE_v10006100mg [Citrus clementina] |
| 7 |
Hb_000120_190 |
0.2396003308 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |
| 8 |
Hb_001260_030 |
0.2431774589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011037_020 |
0.2462441212 |
- |
- |
unknown [Picea sitchensis] |
| 10 |
Hb_000436_080 |
0.2514501957 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_000342_160 |
0.2526205379 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 12 |
Hb_011214_100 |
0.2561261115 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 13 |
Hb_005107_020 |
0.2570050334 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 14 |
Hb_000086_190 |
0.2585674606 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |
| 15 |
Hb_002078_500 |
0.2611868532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002482_060 |
0.2619452503 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |
| 17 |
Hb_000789_150 |
0.2620765616 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000671_120 |
0.2625997712 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 19 |
Hb_003371_170 |
0.264843073 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001396_250 |
0.2651330461 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |