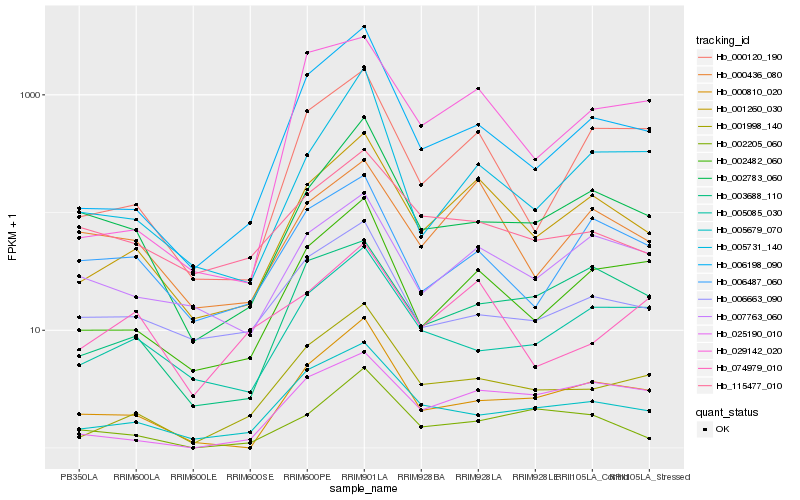
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001260_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002482_060 |
0.1383360414 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |
| 3 |
Hb_000436_080 |
0.1448977289 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_000120_190 |
0.1481371791 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |
| 5 |
Hb_007763_060 |
0.1533169733 |
- |
- |
BnaA02g23510D [Brassica napus] |
| 6 |
Hb_001998_140 |
0.1656367714 |
- |
- |
- |
| 7 |
Hb_006198_090 |
0.168350232 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 8 |
Hb_005679_070 |
0.1703601488 |
- |
- |
hypothetical protein M569_07741, partial [Genlisea aurea] |
| 9 |
Hb_003688_110 |
0.174096416 |
- |
- |
- |
| 10 |
Hb_000810_020 |
0.1751159036 |
- |
- |
- |
| 11 |
Hb_002783_060 |
0.1788021322 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 12 |
Hb_025190_010 |
0.1837641308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006487_060 |
0.1843213443 |
- |
- |
Defective in cullin neddylation protein, putative [Ricinus communis] |
| 14 |
Hb_029142_020 |
0.188600447 |
- |
- |
hypothetical protein POPTR_0004s23900g [Populus trichocarpa] |
| 15 |
Hb_005731_140 |
0.1887968616 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 16 |
Hb_005085_030 |
0.1894420617 |
- |
- |
PREDICTED: N-acetyltransferase 9-like protein isoform X1 [Jatropha curcas] |
| 17 |
Hb_006663_090 |
0.1951533199 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 18 |
Hb_115477_010 |
0.1967425554 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 19 |
Hb_002205_060 |
0.1981972449 |
- |
- |
- |
| 20 |
Hb_074979_010 |
0.2030682301 |
- |
- |
hypothetical protein AALP_AA2G213100 [Arabis alpina] |