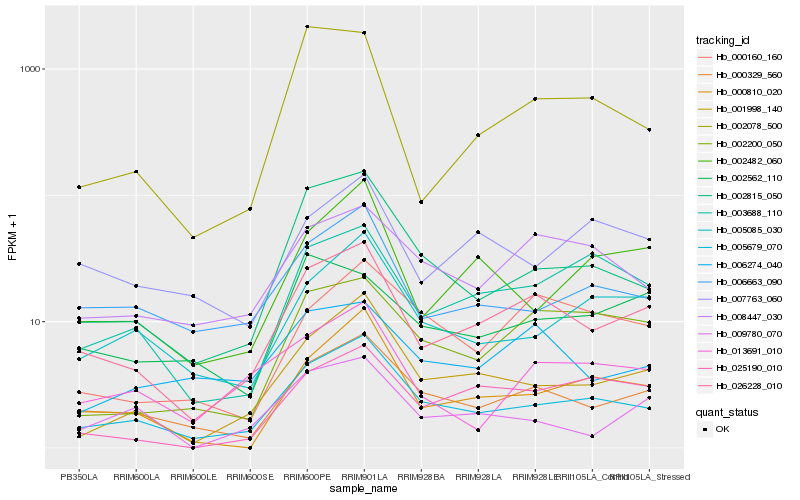
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026228_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000329_560 |
0.1243256339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005679_070 |
0.1376736799 |
- |
- |
hypothetical protein M569_07741, partial [Genlisea aurea] |
| 4 |
Hb_001998_140 |
0.1618394386 |
- |
- |
- |
| 5 |
Hb_003688_110 |
0.1642226815 |
- |
- |
- |
| 6 |
Hb_002200_050 |
0.1733511793 |
- |
- |
unknown [Lotus japonicus] |
| 7 |
Hb_000810_020 |
0.1753975856 |
- |
- |
- |
| 8 |
Hb_025190_010 |
0.175686001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006663_090 |
0.1781111921 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 10 |
Hb_008447_030 |
0.1788565011 |
- |
- |
- |
| 11 |
Hb_002815_050 |
0.1789182726 |
- |
- |
PREDICTED: succinate-semialdehyde dehydrogenase, mitochondrial [Populus euphratica] |
| 12 |
Hb_002482_060 |
0.1797856153 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |
| 13 |
Hb_002078_500 |
0.180328775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006274_040 |
0.183949115 |
- |
- |
PREDICTED: DENN domain and WD repeat-containing protein SCD1-like [Fragaria vesca subsp. vesca] |
| 15 |
Hb_005085_030 |
0.1872995217 |
- |
- |
PREDICTED: N-acetyltransferase 9-like protein isoform X1 [Jatropha curcas] |
| 16 |
Hb_007763_060 |
0.1941940829 |
- |
- |
BnaA02g23510D [Brassica napus] |
| 17 |
Hb_000160_160 |
0.1952452761 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002562_110 |
0.1957047572 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 19 |
Hb_013691_010 |
0.1957182133 |
- |
- |
PREDICTED: uncharacterized protein LOC103724099, partial [Phoenix dactylifera] |
| 20 |
Hb_009780_070 |
0.1961597206 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1 [Jatropha curcas] |